7QPO
 
 | |
6T3E
 
 | | Structure of Thermococcus litoralis Delta(1)-pyrroline-2-carboxylate reductase in complex with NADH and L-proline | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DELTA1-pyrroline-2-carboxylate reductase, PROLINE | | Authors: | Ferraris, D.M, Miggiano, R, Ferrario, E, Rizzi, M. | | Deposit date: | 2019-10-10 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Thermococcus litoralis Delta1-pyrroline-2-carboxylate reductase in complex with NADH and L-proline.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8R15
 
 | | Crystal structure of Fusarium oxysporum NADase I | | Descriptor: | GLYCEROL, TNT domain-containing protein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kallio, J.P, Ferrario, E, Stromland, O, Ziegler, M. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evolution of fungal tuberculosis necrotizing toxin (TNT) domain-containing enzymes reveals divergent adaptations to enhance NAD cleavage.
Protein Sci., 33, 2024
|
|
8R17
 
 | | Crystal structure of Neurospora crassa NADase with modified C-terminus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Conidial surface nicotinamide adenine dinucleotide glycohydrolase,Conidial surface nicotinamide adenine dinucleotide glycohydrolase nadA, ... | | Authors: | Kallio, J.P, Ferrario, E, Ziegler, M. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of fungal tuberculosis necrotizing toxin (TNT) domain-containing enzymes reveals divergent adaptations to enhance NAD cleavage.
Protein Sci., 33, 2024
|
|
8PMR
 
 | | NADase from Aspergillus fumigatus with mutated calcium binding motif (D219A/E220A) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kallio, J.P, Ferrario, E, Stromland, O, Ziegler, M. | | Deposit date: | 2023-06-29 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Novel Calcium-Binding Motif Stabilizes and Increases the Activity of Aspergillus fumigatus Ecto-NADase.
Biochemistry, 62, 2023
|
|
8PMS
 
 | | NADase from Aspergillus fumigatus with replaced C-terminus from Neurospora crassa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kallio, J.P, Ferrario, E, Stromland, O, Ziegler, M. | | Deposit date: | 2023-06-29 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel Calcium-Binding Motif Stabilizes and Increases the Activity of Aspergillus fumigatus Ecto-NADase.
Biochemistry, 62, 2023
|
|
1DF3
 
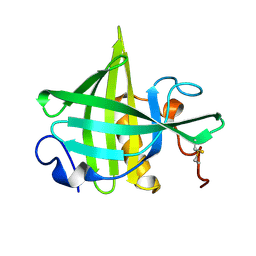 | | SOLUTION STRUCTURE OF A RECOMBINANT MOUSE MAJOR URINARY PROTEIN | | Descriptor: | MAJOR URINARY PROTEIN | | Authors: | Luecke, C, Franzoni, L, Abbate, F, Loehr, F, Ferrari, E, Sorbi, R.T, Rueterjans, H, Spisni, A. | | Deposit date: | 1999-11-17 | | Release date: | 2000-05-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a recombinant mouse major urinary protein.
Eur.J.Biochem., 266, 1999
|
|
1C2P
 
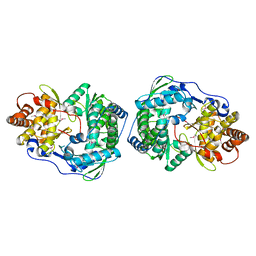 | | HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Descriptor: | RNA-DEPENDENT RNA POLYMERASE | | Authors: | Lesburg, C.A, Cable, M.B, Ferrari, E, Hong, Z, Mannarino, A.F, Weber, P.C. | | Deposit date: | 1999-07-26 | | Release date: | 2000-04-05 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site.
Nat.Struct.Biol., 6, 1999
|
|
2BIC
 
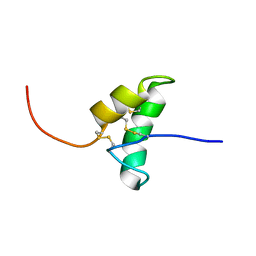 | | The solution structure of the recombinant elicitor protein PcF from the oomycete pathogen P. cactorum | | Descriptor: | PHYTOTOXIC PROTEIN PCF | | Authors: | Nicastro, G, Orsomando, G, Desario, F, Ferrari, E, Manconi, L, Spisni, A, Ruggieri, S. | | Deposit date: | 2005-01-20 | | Release date: | 2006-06-28 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Phytotoxic Protein Pcf: The First Characterized Member of the Phytophthora Pcf Toxin Family.
Protein Sci., 18, 2009
|
|
1JV4
 
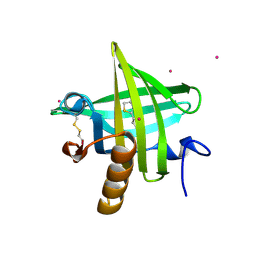 | | Crystal structure of recombinant major mouse urinary protein (rmup) at 1.75 A resolution | | Descriptor: | 2-(SEC-BUTYL)THIAZOLE, CADMIUM ION, Major urinary protein 2 | | Authors: | Kuser, P.R, Franzoni, L, Ferrari, E, Spisni, A, Polikarpov, I. | | Deposit date: | 2001-08-28 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-ray structure of a recombinant major urinary protein at 1.75 A resolution. A comparative study of X-ray and NMR-derived structures.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2K6L
 
 | | The solution structure of XACb0070 from Xanthonomas axonopodis pv citri reveals this new protein is a member of the RHH family of transcriptional repressors | | Descriptor: | Putative uncharacterized protein | | Authors: | Gallo, M, Cicero, D.O, Amata, I, Eliseo, T, Paci, M, Spisni, A, Ferrari, E, Pertinhez, T.A, Farah, C.S. | | Deposit date: | 2008-07-11 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure reveals that XACb0070 from the plant pathogen Xanthomonas citri belongs to the RHH superfamily of bacterial DNA-binding proteins
To be Published
|
|
4MZ4
 
 | | Discovery of an Irreversible HCV NS5B Polymerase Inhibitor | | Descriptor: | 1-[(2-chloroquinolin-3-yl)methyl]-6-fluoro-5-methyl-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxylic acid, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Zeng, Q, Anilkumar, G.N, Rosenblum, S.B, Huang, H.-C, Lesburg, C.A, Jiang, Y, Selyutin, O, Chan, T.-Y, Bennett, F, Chen, K.X, Venkatraman, S, Sannigrahi, M, Velazquez, F, Duca, J.S, Gavalas, S, Huang, Y, Pu, H, Wang, L, Pinto, P, Vibulbhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Li, C, Hesk, D, Gesell, J, Sorota, S, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2013-09-29 | | Release date: | 2013-12-11 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery of an irreversible HCV NS5B polymerase inhibitor.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3U4R
 
 | | Novel HCV NS5B polymerase Inhibitors: Discovery of Indole C2 Acyl sulfonamides | | Descriptor: | 1-[(2-aminopyridin-4-yl)methyl]-5-chloro-N-({3-[(methylsulfonyl)amino]phenyl}sulfonyl)-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxamide, RNA-directed RNA polymerase | | Authors: | Anilkumar, G.N, Selyutin, O, Rosenblum, S.B, Zeng, Q, Jiang, Y, Chan, T.-Y, Pu, H, Wang, L, Bennett, F, Chen, K.X, Lesburg, C.A, Duca, J.S, Gavalas, S, Huang, Y, Pinto, P, Sannagrahi, M, Velazquez, F, Venkataraman, S, Vilbubhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Huang, H.-C, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2011-10-10 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | II. Novel HCV NS5B polymerase inhibitors: Discovery of indole C2 acyl sulfonamides.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3U4O
 
 | | Novel HCV NS5B polymerase Inhibitors: Discovery of Indole C2 Acyl sulfonamides | | Descriptor: | 1-[(2-aminopyridin-4-yl)methyl]-5-chloro-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxylic acid, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Anilkumar, G.N, Selyutin, O, Rosenblum, S.B, Zeng, Q, Jiang, Y, Chan, T.-Y, Pu, H, Wang, L, Bennett, F, Chen, K.X, Lesburg, C.A, Duca, J.S, Gavalas, S, Huang, Y, Pinto, P, Sannigrahi, M, Velazquez, F, Venkataraman, S, Vilbubhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Huang, H.-C, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2011-10-10 | | Release date: | 2011-12-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | II. Novel HCV NS5B polymerase inhibitors: Discovery of indole C2 acyl sulfonamides.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1ST2
 
 | |
2ST1
 
 | |
3LKH
 
 | |
3SKH
 
 | |
3SKE
 
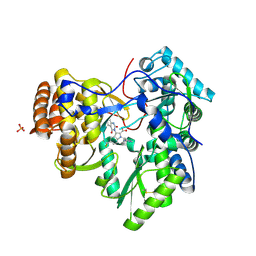 | | I. Novel HCV NS5B Polymerase Inhibitors: Discovery of Indole 2- Carboxylic Acids with C3-Heterocycles | | Descriptor: | 1-[(2-aminopyridin-4-yl)methyl]-3-(2,4-dioxo-1,2-dihydrothieno[3,4-d]pyrimidin-3(4H)-yl)-5-(trifluoromethyl)-1H-indole-2-carboxylic acid, HCV NS5B RNA_DEPENDENT RNA POLYMERASE, PHOSPHATE ION | | Authors: | Lesburg, C.A, Anilkumar, G.N. | | Deposit date: | 2011-06-22 | | Release date: | 2011-08-31 | | Last modified: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | I. Novel HCV NS5B polymerase inhibitors: discovery of indole 2-carboxylic acids with C3-heterocycles.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SKA
 
 | |
3U99
 
 | |
