8G57
 
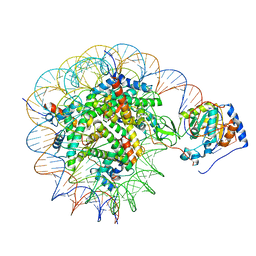 | | Structure of nucleosome-bound Sirtuin 6 deacetylase | | Descriptor: | DNA strand 1, DNA strand 2, Histone H2A type 1-B/E, ... | | Authors: | Chio, U.S, Rechiche, O, Bryll, A.R, Zhu, J, Feldman, J.L, Peterson, C.L, Tan, S, Armache, J.-P. | | Deposit date: | 2023-02-11 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Cryo-EM structure of the human Sirtuin 6-nucleosome complex.
Sci Adv, 9, 2023
|
|
4Y6Q
 
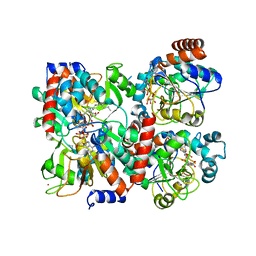 | | Human SIRT2 in complex with 2-O-myristoyl-ADP-ribose | | Descriptor: | NAD-dependent protein deacetylase sirtuin-2, ZINC ION, [(2S,3R,4R,5R)-5-[[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-2,4-bis(oxidanyl)oxolan-3-yl] tetradecanoate | | Authors: | Kudo, N, Ito, A, Yoshida, M. | | Deposit date: | 2015-02-13 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and Structural Basis for Acyl-Group Selectivity and NAD(+) Dependence in Sirtuin-Catalyzed Deacylation.
Biochemistry, 54, 2015
|
|
4Y6O
 
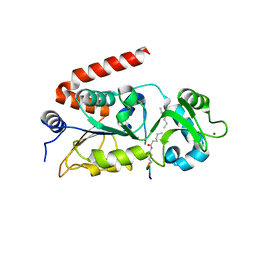 | |
4Y6L
 
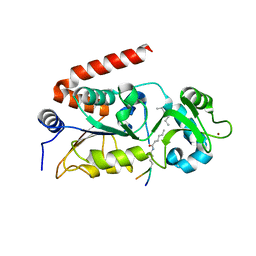 | | Human SIRT2 in complex with myristoylated peptide (H3K9myr) | | Descriptor: | NAD-dependent protein deacetylase sirtuin-2, ZINC ION, peptide THR-ALA-ARG-MYK-SER-THR-GLY | | Authors: | Kudo, N, Ito, A, Yoshida, M. | | Deposit date: | 2015-02-13 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Kinetic and Structural Basis for Acyl-Group Selectivity and NAD(+) Dependence in Sirtuin-Catalyzed Deacylation.
Biochemistry, 54, 2015
|
|
3K35
 
 | | Crystal Structure of Human SIRT6 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent deacetylase sirtuin-6, SULFATE ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-01 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
4ND7
 
 | | Crystal structure of apo 3-nitro-tyrosine tRNA synthetase (5B) in the closed form | | Descriptor: | BETA-MERCAPTOETHANOL, SODIUM ION, Tyrosine--tRNA ligase | | Authors: | Cooley, R.B, Driggers, C.M, Karplus, P.A, Mehl, R.A. | | Deposit date: | 2013-10-25 | | Release date: | 2014-03-19 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Improved Second-Generation 3-Nitro-tyrosine tRNA Synthetases.
Biochemistry, 53, 2014
|
|
4NDA
 
 | | Crystal structure of 3-nitro-tyrosine tRNA synthetase (5B) bound to 3-nitro-tyrosine | | Descriptor: | GLYCEROL, META-NITRO-TYROSINE, SODIUM ION, ... | | Authors: | Cooley, R.B, Driggers, C.M, Karplus, P.A, Mehl, R.A. | | Deposit date: | 2013-10-25 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Improved Second-Generation 3-Nitro-tyrosine tRNA Synthetases.
Biochemistry, 53, 2014
|
|
4ND6
 
 | | Crystal structure of apo 3-nitro-tyrosine tRNA synthetase (5B) in the open form | | Descriptor: | GLYCEROL, Tyrosine-tRNA ligase | | Authors: | Cooley, R.B, Driggers, C.M, Karplus, P.A, Mehl, R.A. | | Deposit date: | 2013-10-25 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Improved Second-Generation 3-Nitro-tyrosine tRNA Synthetases.
Biochemistry, 53, 2014
|
|
3PKJ
 
 | | Human SIRT6 crystal structure in complex with 2'-N-Acetyl ADP ribose | | Descriptor: | NAD-dependent deacetylase sirtuin-6, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Walker, J.R, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
3PKI
 
 | | Human SIRT6 crystal structure in complex with ADP ribose | | Descriptor: | NAD-dependent deacetylase sirtuin-6, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-11 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
