7UFG
 
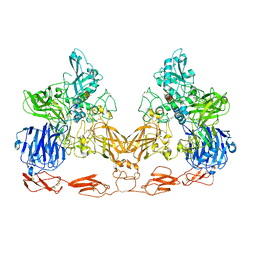 | | Cryo-EM structure of PAPP-A in complex with IGFBP5 | | Descriptor: | Insulin-like growth factor-binding protein 5, Pappalysin-1, ZINC ION | | Authors: | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | Deposit date: | 2022-03-22 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
2AW2
 
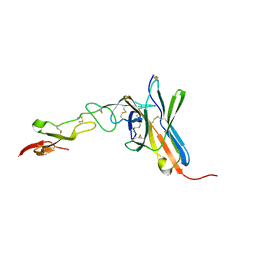 | | Crystal structure of the human BTLA-HVEM complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B and T lymphocyte attenuator, NICKEL (II) ION, ... | | Authors: | Compaan, D.M, Gonzalez, L.C, Tom, I, Loyet, K.M, Eaton, D, Hymowitz, S.G. | | Deposit date: | 2005-08-31 | | Release date: | 2005-09-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Attenuating Lymphocyte Activity: the crystal structure of the BTLA-HVEM complex
J.Biol.Chem., 280, 2005
|
|
8D8O
 
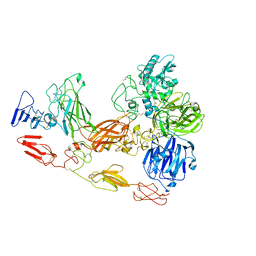 | | Cryo-EM structure of substrate unbound PAPP-A | | Descriptor: | Pappalysin-1, ZINC ION | | Authors: | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | Deposit date: | 2022-06-08 | | Release date: | 2022-09-28 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
8SL1
 
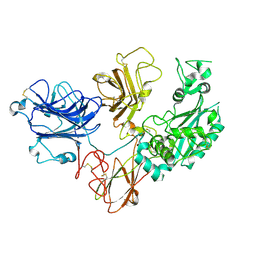 | | Cryo-EM structure of PAPP-A2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Judge, R.A, Stoll, V.S, Eaton, D, Hao, Q, Bratkowski, M.A. | | Deposit date: | 2023-04-20 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Cryo-EM structure of human PAPP-A2 and mechanism of substrate recognition.
Commun Chem, 6, 2023
|
|
8VU5
 
 | | Cryo-EM structure of MPL bound to TPO | | Descriptor: | Thrombopoietin, Thrombopoietin receptor | | Authors: | Bratkowski, M, Hao, Q, Paddock, M. | | Deposit date: | 2024-01-28 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis of MPL activation by thrombopoietin
Blood Vessels, Thrombosis & Hemostasis, 1, 2024
|
|
8GCR
 
 | | HPV16 E6-E6AP-p53 complex | | Descriptor: | Cellular tumor antigen p53, Maltose/maltodextrin-binding periplasmic protein,Protein E6, Ubiquitin-protein ligase E3A, ... | | Authors: | Bratkowski, M.A, Wang, J.C.K, Hao, Q, Nile, A.H. | | Deposit date: | 2023-03-02 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structure of the p53 degradation complex from HPV16.
Nat Commun, 15, 2024
|
|
8U0H
 
 | | Crystal structure of PTPN2 with a PROTAC | | Descriptor: | (5P)-3-(carboxymethoxy)-4-chloro-5-(3-{[(4S)-1-({3-[2-(4-{3-[(3R)-2,6-dioxopiperidin-3-yl]-2-oxo-2,3-dihydro-1,3-benzoxazol-6-yl}piperidin-1-yl)acetamido]phenyl}methanesulfonyl)-2,2-dimethylpiperidin-4-yl]amino}phenyl)thiophene-2-carboxylic acid, ACETATE ION, PTPN2, ... | | Authors: | Jain, R, Longenecker, K, Qiu, W. | | Deposit date: | 2023-08-29 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanistic insights into a heterobifunctional degrader-induced PTPN2/N1 complex.
Commun Chem, 7, 2024
|
|
8UH6
 
 | | Degrader-induced complex between PTPN2 and CRBN-DDB1 | | Descriptor: | (5P)-3-(carboxymethoxy)-4-chloro-5-(3-{[(4S)-1-({3-[(4-{1-[(3R)-2,6-dioxopiperidin-3-yl]-3-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl}piperidine-1-carbonyl)amino]phenyl}methanesulfonyl)-2,2-dimethylpiperidin-4-yl]amino}phenyl)thiophene-2-carboxylic acid, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Catalano, C, Bratkowski, M, Scapin, G, Hao, Q. | | Deposit date: | 2023-10-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanistic insights into a heterobifunctional degrader-induced PTPN2/N1 complex.
Commun Chem, 7, 2024
|
|
7KMA
 
 | | Crystal structure of eif2Balpha with a ligand. | | Descriptor: | 6-O-phosphono-alpha-D-mannopyranose, ACETATE ION, Translation initiation factor eIF-2B subunit alpha | | Authors: | Nocek, B, Hao, Q, Remarcik, C, Stoll, V, Wong, Y, Sidrauski, C. | | Deposit date: | 2020-11-02 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sugar phosphate activation of the stress sensor eIF2B.
Nat Commun, 12, 2021
|
|
7KMF
 
 | | Sugar phosphate activation of the stress sensor eIF2B | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Nocek, B, Hao, Q, Wong, Y, Stoll, V, Sidrauski, C. | | Deposit date: | 2020-11-02 | | Release date: | 2021-07-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Sugar phosphate activation of the stress sensor eIF2B.
Nat Commun, 12, 2021
|
|
4XJS
 
 | | Human CD38 complexed with inhibitor 1 [6-fluoro-2-methyl-4-[(2,3,6-trichlorobenzyl)amino]quinoline-8-carboxamide] | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, 6-fluoro-2-methyl-4-[(2,3,6-trichlorobenzyl)amino]quinoline-8-carboxamide, ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1 | | Authors: | Shewchuk, L.M, Deaton, D, Stewart, E. | | Deposit date: | 2015-01-09 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 4-Amino-8-quinoline Carboxamides as Novel, Submicromolar Inhibitors of NAD-Hydrolyzing Enzyme CD38.
J.Med.Chem., 58, 2015
|
|
