1E0V
 
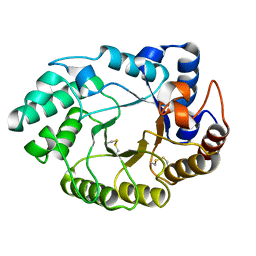 | | Xylanase 10A from Sreptomyces lividans. cellobiosyl-enzyme intermediate at 1.7 A | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
1ESC
 
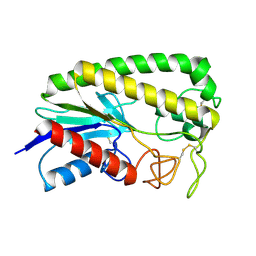 | | THE MOLECULAR MECHANISM OF ENANTIORECOGNITION BY ESTERASES | | Descriptor: | ESTERASE | | Authors: | Wei, Y, Schottel, J.L, Derewenda, U, Swenson, L, Patkar, S, Derewenda, Z.S. | | Deposit date: | 1994-10-07 | | Release date: | 1995-10-15 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel variant of the catalytic triad in the Streptomyces scabies esterase.
Nat.Struct.Biol., 2, 1995
|
|
1F7C
 
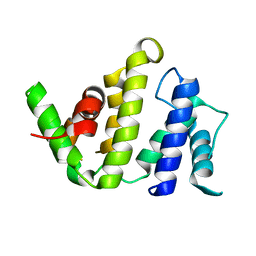 | | CRYSTAL STRUCTURE OF THE BH DOMAIN FROM GRAF, THE GTPASE REGULATOR ASSOCIATED WITH FOCAL ADHESION KINASE | | Descriptor: | RHOGAP PROTEIN | | Authors: | Longenecker, K.L, Derewenda, U, Sheffield, P.J, Zheng, Y, Derewenda, Z.S. | | Deposit date: | 2000-06-26 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the BH domain from graf and its implications for Rho GTPase recognition.
J.Biol.Chem., 275, 2000
|
|
2BTN
 
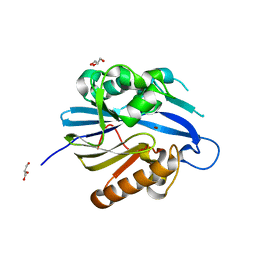 | | Crystal Structure and Catalytic Mechanism of the Quorum-Quenching N- Acyl Homoserine Lactone Hydrolase | | Descriptor: | AIIA-LIKE PROTEIN, GLYCEROL, ZINC ION | | Authors: | Kim, M.H, Choi, W.C, Kang, H.O, Lee, J.S, Kang, B.S, Kim, K.J, Derewenda, Z.S, Oh, T.K, Lee, C.H, Lee, J.K. | | Deposit date: | 2005-06-03 | | Release date: | 2005-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Molecular Structure and Catalytic Mechanism of a Quorum-Quenching N-Acyl-L-Homoserine Lactone Hydrolase.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BR6
 
 | | Crystal Structure of Quorum-Quenching N-Acyl Homoserine Lactone Lactonase | | Descriptor: | AIIA-LIKE PROTEIN, GLYCEROL, HOMOSERINE LACTONE, ... | | Authors: | Kim, M.H, Choi, W.C, Kang, H.O, Kang, B.S, Kim, K.J, Derewenda, Z.S, Lee, J.K, Oh, T.K, Lee, C.H. | | Deposit date: | 2005-05-03 | | Release date: | 2005-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Molecular Structure and Catalytic Mechanism of a Quorum-Quenching N-Acyl-L-Homoserine Lactone Hydrolase.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BXW
 
 | | CRYSTAL STRUCTURE OF RHOGDI Lys(135,138,141)Tyr MUTANT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, RHO GDP-DISSOCIATION INHIBITOR 1 | | Authors: | Sikorska, M, Cooper, D.R, Otlewski, J, Derewenda, Z.S. | | Deposit date: | 2005-07-27 | | Release date: | 2005-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein Crystallization by Surface Entropy Reduction: Optimization of the Ser Strategy
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2BJO
 
 | | Crystal Structure of the Organic Hydroperoxide Resistance Protein OhrB of Bacillus subtilis | | Descriptor: | ORGANIC HYDROPEROXIDE RESISTANCE PROTEIN OHRB | | Authors: | Cooper, D.R, Surendranath, Y, Bielnicki, J, Devedjiev, Y, Joachimiak, A, Derewenda, Z.S. | | Deposit date: | 2005-02-04 | | Release date: | 2006-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Bacillus Subtilis Ohrb Hydroperoxide-Resistance Protein in a Fully Oxidized State.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3N0U
 
 | |
1TIA
 
 | | AN UNUSUAL BURIED POLAR CLUSTER IN A FAMILY OF FUNGAL LIPASES | | Descriptor: | LIPASE | | Authors: | Derewenda, U, Swenson, L, Yamaguchi, S, Wei, Y, Derewenda, Z.S. | | Deposit date: | 1993-12-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An unusual buried polar cluster in a family of fungal lipases.
Nat.Struct.Biol., 1, 1994
|
|
1JKM
 
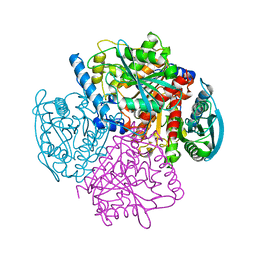 | | BREFELDIN A ESTERASE, A BACTERIAL HOMOLOGUE OF HUMAN HORMONE SENSITIVE LIPASE | | Descriptor: | BREFELDIN A ESTERASE | | Authors: | Wei, Y, Contreras, A.J, Sheffield, P, Osterlund, T, Derewenda, U, Matern, U.O, Derewenda, Z.S. | | Deposit date: | 1998-02-04 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of brefeldin A esterase, a bacterial homolog of the mammalian hormone-sensitive lipase.
Nat.Struct.Biol., 6, 1999
|
|
1KLL
 
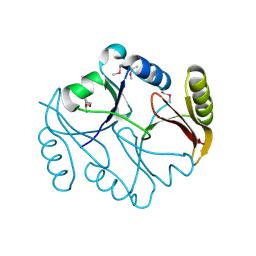 | | Molecular basis of mitomycin C resictance in streptomyces: Crystal structures of the MRD protein with and without a drug derivative | | Descriptor: | 1,2-CIS-1-HYDROXY-2,7-DIAMINO-MITOSENE, mitomycin-binding protein | | Authors: | Martin, T.W, Dauter, Z, Devedjiev, Y, Sheffield, P, Jelen, F, He, M, Sherman, D, Otlewski, J, Derewenda, Z.S, Derewenda, U. | | Deposit date: | 2001-12-12 | | Release date: | 2002-07-19 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
Structure, 10, 2002
|
|
1KMZ
 
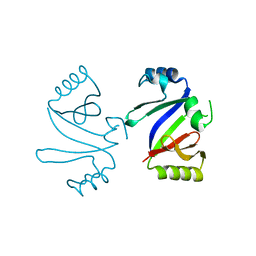 | | MOLECULAR BASIS OF MITOMYCIN C RESICTANCE IN STREPTOMYCES: CRYSTAL STRUCTURES OF THE MRD PROTEIN WITH AND WITHOUT A DRUG DERIVATIVE | | Descriptor: | mitomycin-binding protein | | Authors: | Martin, T.W, Dauter, Z, Devedjiev, Y, Sheffield, P, Jelen, F, He, M, Sherman, D, Otlewski, J, Derewenda, Z.S, Derewenda, U. | | Deposit date: | 2001-12-17 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
Structure, 10, 2002
|
|
3E57
 
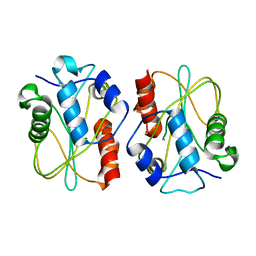 | |
1ES9
 
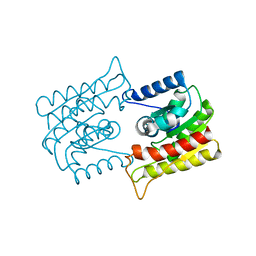 | | X-RAY CRYSTAL STRUCTURE OF R22K MUTANT OF THE MAMMALIAN BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASES (PAF-AH) | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE IB GAMMA SUBUNIT | | Authors: | McMullen, T.W.P, Li, J, Sheffield, P.J, Aoki, J, Martin, T.W, Arai, H, Inoue, K, Derewenda, Z.S. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The functional implications of the dimerization of the catalytic subunits of the mammalian brain platelet-activating factor acetylhydrolase (Ib).
Protein Eng., 13, 2000
|
|
1H4R
 
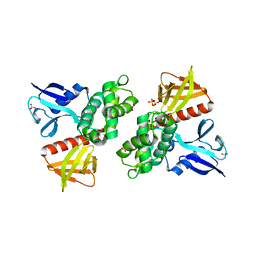 | | Crystal Structure of the FERM domain of Merlin, the Neurofibromatosis 2 Tumor Suppressor Protein. | | Descriptor: | MERLIN, SULFATE ION | | Authors: | Cooper, D.R, Kang, B.S, Sheffield, P, Devedjiev, Y, Derewenda, Z.S. | | Deposit date: | 2001-05-14 | | Release date: | 2002-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of the Ferm Domain of Merlin, the Neurofibromatosis Type 2 Gene Product.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1JFR
 
 | |
1ESE
 
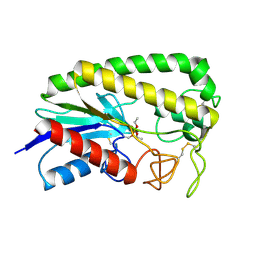 | | THE MOLECULAR MECHANISM OF ENANTIORECOGNITION BY ESTERASES | | Descriptor: | DIETHYL PHOSPHONATE, ESTERASE | | Authors: | Wei, Y, Schottel, J.L, Derewenda, U, Swenson, L, Patkar, S, Derewenda, Z.S. | | Deposit date: | 1994-10-07 | | Release date: | 1995-10-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel variant of the catalytic triad in the Streptomyces scabies esterase.
Nat.Struct.Biol., 2, 1995
|
|
1ESD
 
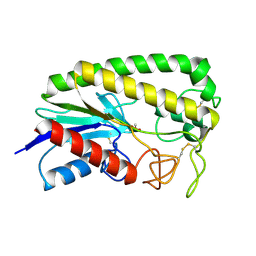 | | THE MOLECULAR MECHANISM OF ENANTIORECOGNITION BY ESTERASES | | Descriptor: | ESTERASE, METHYLPHOSPHONIC ACID ESTER GROUP | | Authors: | Wei, Y, Schottel, J.L, Derewenda, U, Swenson, L, Patkar, S, Derewenda, Z.S. | | Deposit date: | 1994-10-07 | | Release date: | 1995-10-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel variant of the catalytic triad in the Streptomyces scabies esterase.
Nat.Struct.Biol., 2, 1995
|
|
1FTN
 
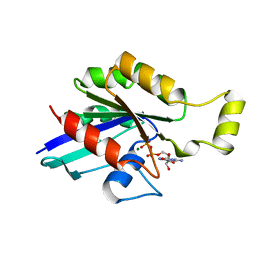 | | CRYSTAL STRUCTURE OF THE HUMAN RHOA/GDP COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, TRANSFORMING PROTEIN RHOA (H12) | | Authors: | Wei, Y, Zhang, Y, Derewenda, U, Liu, X, Minor, W, Nakamoto, R.K, Somlyo, A.V, Somlyo, A.P, Derewenda, Z.S. | | Deposit date: | 1997-03-13 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of RhoA-GDP and its functional implications.
Nat.Struct.Biol., 4, 1997
|
|
2JHX
 
 | |
2JHV
 
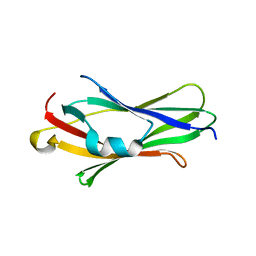 | |
2JHU
 
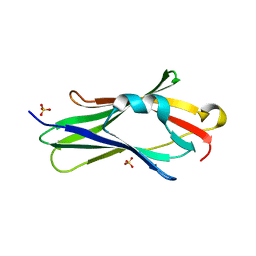 | | CRYSTAL STRUCTURE OF RHOGDI E154A,E155A MUTANT | | Descriptor: | RHO GDP-DISSOCIATION INHIBITOR 1, SULFATE ION | | Authors: | Cooper, D.R, Pinkowska, M, Derewenda, Z.S. | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Protein Crystallization by Surface Entropy Reduction: Optimization of the Ser Strategy
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2JI0
 
 | | CRYSTAL STRUCTURE OF RHOGDI K138Y, K141Y MUTANT | | Descriptor: | RHO GDP-DISSOCIATION INHIBITOR 1, SULFATE ION | | Authors: | Cooper, D.R, Boczek, T, Derewenda, Z.S. | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein Crystallization by Surface Entropy Reduction: Optimization of the Ser Strategy
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2JHT
 
 | | CRYSTAL STRUCTURE OF RHOGDI K135T,K138T,K141T MUTANT | | Descriptor: | LITHIUM ION, RHO GDP-DISSOCIATION INHIBITOR 1, SULFATE ION | | Authors: | Cooper, D.R, Grelewska, K, Derewenda, Z.S. | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Protein Crystallization by Surface Entropy Reduction: Optimization of the Ser Strategy
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2JHW
 
 | | CRYSTAL STRUCTURE OF RHOGDI E155A, E157A MUTANT | | Descriptor: | RHO GDP-DISSOCIATION INHIBITOR 1, SULFATE ION | | Authors: | Cooper, D.R, Grelewska, K, Derewenda, Z.S. | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein Crystallization by Surface Entropy Reduction: Optimization of the Ser Strategy
Acta Crystallogr.,Sect.D, 63, 2007
|
|
