1WNH
 
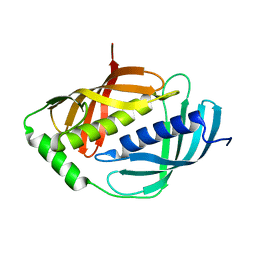 | | Crystal structure of mouse Latexin (tissue carboxypeptidase inhibitor) | | Descriptor: | Latexin | | Authors: | Aagaard, A, Listwan, P, Cowieson, N, Huber, T, Ravasi, T, Wells, C.A, Flanagan, J.U, Hume, D.A, Kobe, B, Martin, J.L. | | Deposit date: | 2004-08-04 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | An Inflammatory Role for the Mammalian Carboxypeptidase Inhibitor Latexin: Relationship to Cystatins and the Tumor Suppressor TIG1
Structure, 13, 2005
|
|
4BVR
 
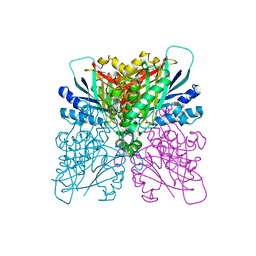 | | Cyanuric acid hydrolase: evolutionary innovation by structural concatenation. | | Descriptor: | 1,3,5-triazine-2,4,6-triol, CYANURIC ACID AMIDOHYDROLASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, French, N.G, Briggs, L.J, Panjikar, S, Cowieson, N, Newman, J, Scott, C. | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Cyanuric Acid Hydrolase: Evolutionary Innovation by Structural Concatenation.
Mol.Microbiol., 88, 2013
|
|
4B2G
 
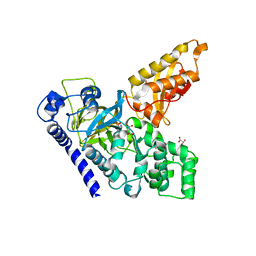 | | Crystal Structure of an Indole-3-Acetic Acid Amido Synthase from Vitis vinifera Involved in Auxin Homeostasis | | Descriptor: | GH3-1 AUXIN CONJUGATING ENZYME, MALONATE ION, [(2S,3R,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl] 2-(1H-indol-3-yl)ethyl hydrogen phosphate | | Authors: | Peat, T.S, Bottcher, C, Newman, J, Lucent, D, Cowieson, N, Davies, C. | | Deposit date: | 2012-07-16 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of an Indole-3-Acetic Acid Amido Synthetase from Grapevine Involved in Auxin Homeostasis.
Plant Cell, 24, 2012
|
|
4CYY
 
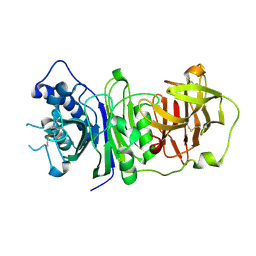 | | The structure of vanin-1: defining the link between metabolic disease, oxidative stress and inflammation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PANTETHEINASE | | Authors: | Boersma, Y.L, Newman, J, Adams, T.E, Sparrow, L, Cowieson, N, Lucent, D, Krippner, G, Bozaoglu, K, Peat, T.S. | | Deposit date: | 2014-04-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | The Structure of Vanin-1: A Key Enzyme Linking Metabolic Disease and Inflammation
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CYG
 
 | | The structure of vanin-1: defining the link between metabolic disease, oxidative stress and inflammation | | Descriptor: | (2R)-2,4-dihydroxy-N-[(3S)-3-hydroxy-4-phenylbutyl]-3,3-dimethylbutanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Boersma, Y.L, Newman, J, Adams, T.E, Sparrow, L, Cowieson, N, Lucent, D, Krippner, G, Bozaoglu, K, Peat, T.S. | | Deposit date: | 2014-04-11 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of Vanin-1: A Key Enzyme Linking Metabolic Disease and Inflammation
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CYF
 
 | | The structure of vanin-1: defining the link between metabolic disease, oxidative stress and inflammation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PANTETHEINASE | | Authors: | Boersma, Y.L, Newman, J, Adams, T.E, Sparrow, L, Cowieson, N, Lucent, D, Krippner, G, Bozaoglu, K, Peat, T.S. | | Deposit date: | 2014-04-11 | | Release date: | 2014-12-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of Vanin-1: A Key Enzyme Linking Metabolic Disease and Inflammation
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4BVS
 
 | | Cyanuric acid hydrolase: evolutionary innovation by structural concatenation. | | Descriptor: | 1,3,5-triazine-2,4,6-triamine, CYANURIC ACID AMIDOHYDROLASE, MAGNESIUM ION | | Authors: | Peat, T.S, Balotra, S, Wilding, M, French, N.G, Briggs, L.J, Panjikar, S, Cowieson, N, Newman, J, Scott, C. | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cyanuric Acid Hydrolase: Evolutionary Innovation by Structural Concatenation.
Mol.Microbiol., 88, 2013
|
|
4BVT
 
 | | Cyanuric acid hydrolase: evolutionary innovation by structural concatenation | | Descriptor: | BARBITURIC ACID, CYANURIC ACID AMIDOHYDROLASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, French, N.G, Briggs, L.J, Panjikar, S, Cowieson, N, Newman, J, Scott, C. | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Cyanuric Acid Hydrolase: Evolutionary Innovation by Structural Concatenation.
Mol.Microbiol., 88, 2013
|
|
4BVQ
 
 | | Cyanuric acid hydrolase: evolutionary innovation by structural concatenation. | | Descriptor: | CYANURIC ACID AMIDOHYDROLASE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Peat, T.S, Balotra, S, Wilding, M, French, N.G, Briggs, L.J, Panjikar, S, Cowieson, N, Newman, J, Scott, C. | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cyanuric Acid Hydrolase: Evolutionary Innovation by Structural Concatenation.
Mol.Microbiol., 88, 2013
|
|
4ZRB
 
 | |
4R4U
 
 | |
4MOB
 
 | | Acyl-Coenzyme A thioesterase 12 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Acyl-coenzyme A thioesterase 12, COENZYME A | | Authors: | Swarbrick, C.M.D, Forwood, J.K. | | Deposit date: | 2013-09-12 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis for regulation of the human acetyl-CoA thioesterase 12 and interactions with the steroidogenic acute regulatory protein-related lipid transfer (START) domain.
J.Biol.Chem., 289, 2014
|
|
4MOC
 
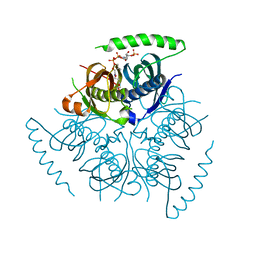 | | Human Acyl-coenzyme A Thioesterase 12 | | Descriptor: | Acyl-coenzyme A thioesterase 12, COENZYME A | | Authors: | Swarbrick, C.M.D, Forwood, J.K. | | Deposit date: | 2013-09-12 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for regulation of the human acetyl-CoA thioesterase 12 and interactions with the steroidogenic acute regulatory protein-related lipid transfer (START) domain.
J.Biol.Chem., 289, 2014
|
|
8PAB
 
 | |
8PAE
 
 | |
8PAG
 
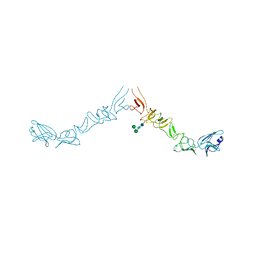 | | Crystal structure of the ectodomain of Norway rat pestivirus E2 glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Aitkenhead, H, Stuart, D.I, EL Omari, K. | | Deposit date: | 2023-06-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural comparison of typical and atypical E2 pestivirus glycoproteins.
Structure, 32, 2024
|
|
8RSI
 
 | | Crystal structure of Methanobrevibacter oralis macrodomain | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTAMIC ACID, ... | | Authors: | Ariza, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Evolutionary and molecular basis of ADP-ribosylation reversal by zinc-dependent macrodomains.
J.Biol.Chem., 2024
|
|
8RSJ
 
 | |
8RSM
 
 | | Crystal structure of Streptococcus pyogenes macrodomain in complex with ADP-ribose | | Descriptor: | Protein-ADP-ribose hydrolase, ZINC ION, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Ariza, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Evolutionary and molecular basis of ADP-ribosylation reversal by zinc-dependent macrodomains.
J.Biol.Chem., 2024
|
|
8RSK
 
 | | Crystal structure of Methanobrevibacter oralis macrodomain in complex with Asn-ADPr | | Descriptor: | (2~{S})-4-[[(2~{S},3~{R},4~{S},5~{R})-5-[[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-3,4-bis(oxidanyl)oxolan-2-yl]amino]-2-azanyl-4-oxidanylidene-butanoic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Ariza, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Evolutionary and molecular basis of ADP-ribosylation reversal by zinc-dependent macrodomains.
J.Biol.Chem., 2024
|
|
8RSN
 
 | |
8RSL
 
 | | Crystal structure of Staphylococcus aureus macrodomain | | Descriptor: | Protein-ADP-ribose hydrolase, ZINC ION | | Authors: | Ariza, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Evolutionary and molecular basis of ADP-ribosylation reversal by zinc-dependent macrodomains.
J.Biol.Chem., 2024
|
|
5T02
 
 | | Structural characterisation of mutant Asp39Ala of thioesterase (NmACH) from Neisseria meningitidis | | Descriptor: | Acyl-CoA hydrolase, CHLORIDE ION, COENZYME A, ... | | Authors: | Khandokar, Y.B, Srivastava, P, Forwood, J.K. | | Deposit date: | 2016-08-15 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into GDP-mediated regulation of a bacterial acyl-CoA thioesterase.
J. Biol. Chem., 292, 2017
|
|
5SZZ
 
 | | Novel Structural Insights into GDP-Mediated Regulation of Acyl-CoA Thioesterases | | Descriptor: | Acyl-CoA hydrolase, CHLORIDE ION, COENZYME A, ... | | Authors: | Khandokar, Y.B, Srivastava, P, Forwood, J.K. | | Deposit date: | 2016-08-15 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into GDP-mediated regulation of a bacterial acyl-CoA thioesterase.
J. Biol. Chem., 292, 2017
|
|
5SZU
 
 | |
