4U7D
 
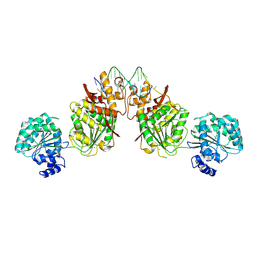 | | Structure of human RECQ-like helicase in complex with an oligonucleotide | | 分子名称: | ATP-dependent DNA helicase Q1, DNA oligonucleotide, ZINC ION | | 著者 | Pike, A.C.W, Zhang, Y, Schnecke, C, Cooper, C.D.O, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | 登録日 | 2014-07-30 | | 公開日 | 2015-01-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Human RECQ1 helicase-driven DNA unwinding, annealing, and branch migration: Insights from DNA complex structures.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5T3A
 
 | |
5LLJ
 
 | |
6YUW
 
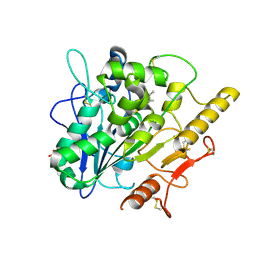 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 454 | | 分子名称: | 1-(cyclopropylmethyl)-2,5-dimethyl-pyrrole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | 著者 | Ruza, R.R, Hillier, J, Jones, E.Y. | | 登録日 | 2020-04-27 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YV2
 
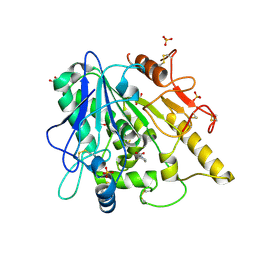 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLIDINE-3-CARBOXYLIC ACID FRAGMENT 598 | | 分子名称: | (3~{R})-1-phenylpyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ruza, R.R, Hillier, J, Jones, E.Y. | | 登録日 | 2020-04-27 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YXI
 
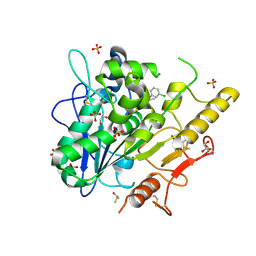 | | Structure of Notum in complex with a 1-(3-Chlorophenyl)-2,5-dimethyl-1H-pyrrole-3-carboxylic acid inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 1-(3-chlorophenyl)-2,5-dimethyl-pyrrole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Vecchia, L, Jones, E.Y, Ruza, R.R, Hillier, J, Zhao, Y. | | 登録日 | 2020-05-01 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YV4
 
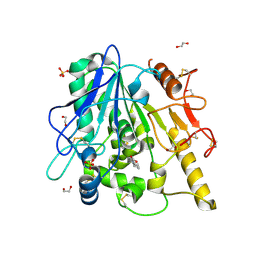 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 686 | | 分子名称: | 1,2-ETHANEDIOL, 1-cyclopropyl-2,5-dimethyl-pyrrole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Hillier, J, Ruza, R.R, Jones, E.Y. | | 登録日 | 2020-04-27 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YV0
 
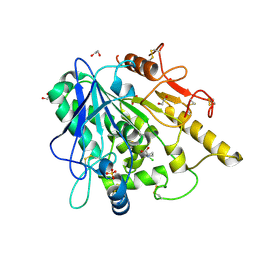 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLIDINE-3-CARBOXYLIC ACID FRAGMENT 587 | | 分子名称: | (3~{R})-1-(2-chlorophenyl)pyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ruza, R.R, Hillier, J, Jones, E.Y. | | 登録日 | 2020-04-27 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YUY
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 471 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-methyl-5-(trifluoromethyl)-1~{H}-pyrrole-3-carboxylic acid, DIMETHYL SULFOXIDE, ... | | 著者 | Hillier, J, Ruza, R.R, Jones, E.Y. | | 登録日 | 2020-04-27 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YSK
 
 | | 1-phenylpyrroles and 1-enylpyrrolidines as inhibitors of Notum | | 分子名称: | (3~{S})-1-[4-chloranyl-3-(trifluoromethyl)phenyl]pyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhao, Y, Jones, E.Y, Fish, P. | | 登録日 | 2020-04-22 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
1EJ0
 
 | | FTSJ RNA METHYLTRANSFERASE COMPLEXED WITH S-ADENOSYLMETHIONINE, MERCURY DERIVATIVE | | 分子名称: | FTSJ, MERCURY (II) ION, S-ADENOSYLMETHIONINE | | 著者 | Bugl, H, Fauman, E.B, Staker, B.L, Zheng, F, Kushner, S.R, Saper, M.A, Bardwell, J.C.A, Jakob, U. | | 登録日 | 2000-02-29 | | 公開日 | 2000-08-30 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | RNA methylation under heat shock control.
Mol.Cell, 6, 2000
|
|
1EIZ
 
 | | FTSJ RNA METHYLTRANSFERASE COMPLEXED WITH S-ADENOSYLMETHIONINE | | 分子名称: | FTSJ, S-ADENOSYLMETHIONINE | | 著者 | Bugl, H, Fauman, E.B, Staker, B.L, Zheng, F, Kushner, S.R, Saper, M.A, Bardwell, J.C.A, Jakob, U. | | 登録日 | 2000-02-29 | | 公開日 | 2000-08-30 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | RNA methylation under heat shock control.
Mol.Cell, 6, 2000
|
|
3F8T
 
 | |
6ZHX
 
 | | Cryo-EM structure of the regulatory linker of ALC1 bound to the nucleosome's acidic patch: nucleosome class. | | 分子名称: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (145-MER) Widom 601 sequence, Histone H2A type 1, ... | | 著者 | Bacic, L, Gaullier, G, Croll, T.I, Deindl, S. | | 登録日 | 2020-06-24 | | 公開日 | 2020-12-23 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch.
Cell Rep, 33, 2020
|
|
6ZHY
 
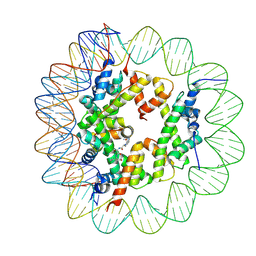 | | Cryo-EM structure of the regulatory linker of ALC1 bound to the nucleosome's acidic patch: hexasome class. | | 分子名称: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (110-MER) Widom 601 sequence, Histone H2A type 1, ... | | 著者 | Bacic, L, Gaullier, G, Deindl, S. | | 登録日 | 2020-06-24 | | 公開日 | 2020-12-23 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch.
Cell Rep, 33, 2020
|
|
4C8H
 
 | |
4C8S
 
 | |
4C95
 
 | |
4C93
 
 | |
3UL5
 
 | | Saccharum officinarum canecystatin-1 in space group C2221 | | 分子名称: | Canecystatin-1, GLYCEROL, SODIUM ION | | 著者 | Valadares, N.F, Pereira, H.M, Oliveira-Silva, R, Garratt, R.C. | | 登録日 | 2011-11-10 | | 公開日 | 2012-11-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | X-ray crystallography and NMR studies of domain-swapped canecystatin-1.
Febs J., 280, 2013
|
|
3UL6
 
 | | Saccharum officinarum canecystatin-1 in space group P6422 | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Canecystatin-1 | | 著者 | Valadares, N.F, Pereira, H.M, Oliveira-Silva, R, Garratt, R.C. | | 登録日 | 2011-11-10 | | 公開日 | 2012-12-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | X-ray crystallography and NMR studies of domain-swapped canecystatin-1.
Febs J., 280, 2013
|
|
5MEC
 
 | |
5MEA
 
 | | Crystal structure of yeast Cdt1 (N terminal and middle domain), form 2. | | 分子名称: | Cell division cycle protein CDT1, GLYCEROL, SULFATE ION | | 著者 | Pye, V.E, Frigola, J, Diffley, J.F.X, Cherepanov, P. | | 登録日 | 2016-11-14 | | 公開日 | 2017-05-17 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.152 Å) | | 主引用文献 | Cdt1 stabilizes an open MCM ring for helicase loading.
Nat Commun, 8, 2017
|
|
5MEB
 
 | | Crystal structure of yeast Cdt1 C-terminal domain | | 分子名称: | Cell division cycle protein CDT1, SULFATE ION | | 著者 | Pye, V.E, Frigola, J, Diffley, J.F.X, Cherepanov, P. | | 登録日 | 2016-11-14 | | 公開日 | 2017-05-17 | | 最終更新日 | 2017-07-05 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Cdt1 stabilizes an open MCM ring for helicase loading.
Nat Commun, 8, 2017
|
|
5ME9
 
 | | Crystal structure of yeast Cdt1 (N terminal and middle domain), form 1. | | 分子名称: | Cell division cycle protein CDT1, GLYCEROL, SULFATE ION | | 著者 | Pye, V.E, Frigola, J, Diffley, J.F.X, Cherepanov, P. | | 登録日 | 2016-11-14 | | 公開日 | 2017-05-17 | | 最終更新日 | 2017-07-05 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Cdt1 stabilizes an open MCM ring for helicase loading.
Nat Commun, 8, 2017
|
|
