1OEW
 
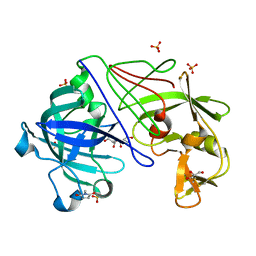 | | ATOMIC RESOLUTION STRUCTURE OF NATIVE ENDOTHIAPEPSIN | | Descriptor: | ENDOTHIAPEPSIN, GLYCEROL, SERINE, ... | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2003-03-31 | | Release date: | 2003-04-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
6T3G
 
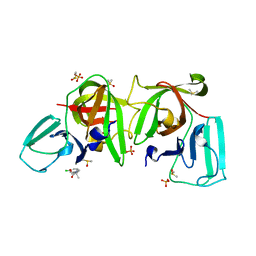 | |
6T4S
 
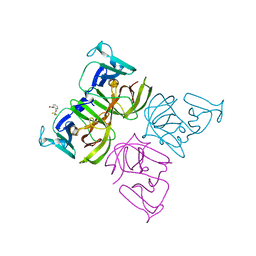 | |
6T5D
 
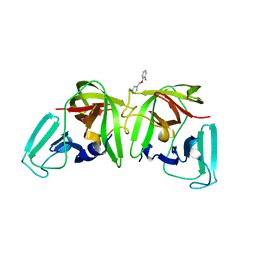 | |
6TAL
 
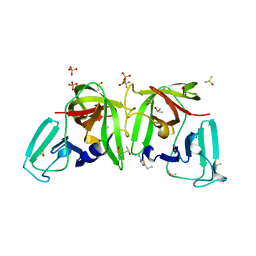 | | 3C-like protease from Southampton virus complexed with FMOPL000227a. | | Descriptor: | 5-ethyl-1,3,4-thiadiazol-2-amine, DIMETHYL SULFOXIDE, Genome polyprotein, ... | | Authors: | Guo, J, Cooper, J.B. | | Deposit date: | 2019-10-29 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | In crystallo-screening for discovery of human norovirus 3C-like protease inhibitors.
J Struct Biol X, 4, 2020
|
|
1QN2
 
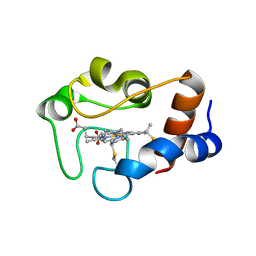 | | cytochrome cH from Methylobacterium extorquens | | Descriptor: | CYTOCHROME CH, HEME C | | Authors: | Read, J, Gill, R, Dales, S.L, Cooper, J.B, Wood, S.P, Anthony, C. | | Deposit date: | 1999-10-13 | | Release date: | 2000-10-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Molecular Structure of an Unusual Cytochrome C2 Determined at 2.0A; the Cytochrome cH from Methylobacterium Extorquens
Protein Sci., 8, 1999
|
|
1EPN
 
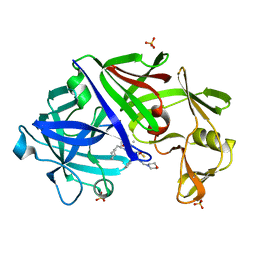 | | A STRUCTURAL COMPARISON OF 21 INHIBITOR COMPLEXES OF THE ASPARTIC PROTEINASE FROM ENDOTHIA PARASITICA | | Descriptor: | ENDOTHIAPEPSIN, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R,2S)-1-(cyclohexylmethyl)-2-hydroxy-3-(1-methylethoxy)-3-oxopropyl]-S-methyl-L-cysteinamide, SULFATE ION | | Authors: | Crawford, M, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
4LP9
 
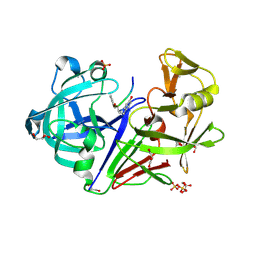 | | Endothiapepsin complexed with Phe-reduced-Tyr peptide. | | Descriptor: | Endothiapepsin, GLYCEROL, SULFATE ION, ... | | Authors: | Guo, J, Cooper, J.B, Wood, S.P. | | Deposit date: | 2013-07-15 | | Release date: | 2014-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structure of endothiapepsin complexed with a Phe-Tyr reduced-bond inhibitor at 1.35 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
1FQ6
 
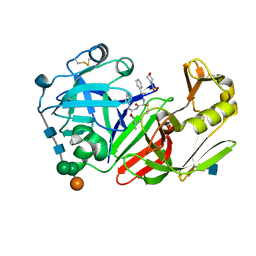 | | X-RAY STRUCTURE OF GLYCOL INHIBITOR PD-133,450 BOUND TO SACCHAROPEPSIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(1S)-2-{[(2S,3R,4S)-1-cyclohexyl-3,4-dihydroxy-6-methylheptan-2-yl]amino}-1-(ethylsulfanyl)-2-oxoethyl]-Nalpha-(morp holin-4-ylsulfonyl)-L-phenylalaninamide, SACCHAROPEPSIN, ... | | Authors: | Cronin, N.B, Badasso, M.O, Tickle, I.J, Dreyer, T, Hoover, D.J, Rosati, R.L, Humblet, C.C, Lunney, E.A, Cooper, J.B. | | Deposit date: | 2000-09-03 | | Release date: | 2000-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray structures of five renin inhibitors bound to saccharopepsin: exploration of active-site specificity.
J.Mol.Biol., 303, 2000
|
|
1FQ7
 
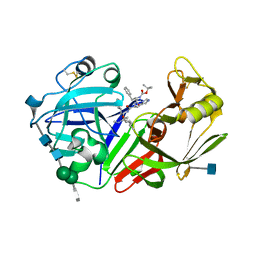 | | X-RAY STRUCTURE OF INHIBITOR CP-72,647 BOUND TO SACCHAROPEPSIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-(tert-butoxycarbonyl)-L-phenylalanyl-N-[(2S,3S,5R)-1-cyclohexyl-3-hydroxy-7-methyl-5-(methylcarbamoyl)octan-2-yl]-L-histidinamide, SACCHAROPEPSIN, ... | | Authors: | Cronin, N.B, Badasso, M.O, Tickle, I.J, Dreyer, T, Hoover, D.J, Rosati, R.L, Humblet, C.C, Lunney, E.A, Cooper, J.B. | | Deposit date: | 2000-09-04 | | Release date: | 2000-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structures of five renin inhibitors bound to saccharopepsin: exploration of active-site specificity.
J.Mol.Biol., 303, 2000
|
|
4MLV
 
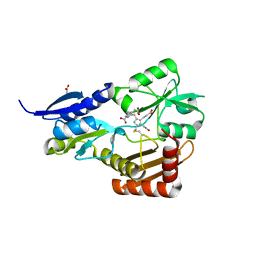 | | Crystal Structure of Bacillus megaterium porphobilinogen deaminase | | Descriptor: | 3-[(5S)-5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]propanoic acid, 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, ACETIC ACID, ... | | Authors: | Azim, N, Deery, E, Warren, M.J, Erskine, P, Cooper, J.B, Coker, A, Wood, S.P, Akhtar, M. | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.455 Å) | | Cite: | Structural evidence for the partially oxidized dipyrromethene and dipyrromethanone forms of the cofactor of porphobilinogen deaminase: structures of the Bacillus megaterium enzyme at near-atomic resolution.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1FQ8
 
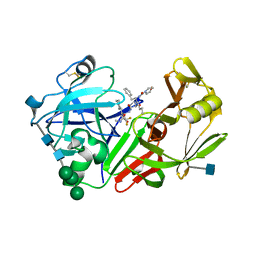 | | X-RAY STRUCTURE OF DIFLUOROSTATINE INHIBITOR CP81,198 BOUND TO SACCHAROPEPSIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(2S)-1-[[(2S)-1-[[(2S,3R)-1-cyclohexyl-4,4-difluoro-3-hydroxy-5-(methylamino)-5-oxo-pentan-2-yl]amino]-1-oxo-hexan-2 -yl]amino]-1-oxo-3-phenyl-propan-2-yl]morpholine-4-carboxamide, SACCHAROPEPSIN, ... | | Authors: | Cronin, N.B, Badasso, M.O, Tickle, I.J, Dreyer, T, Hoover, D.J, Rosati, R.L, Humblet, C.C, Lunney, E.A, Cooper, J.B. | | Deposit date: | 2000-09-04 | | Release date: | 2000-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structures of five renin inhibitors bound to saccharopepsin: exploration of active-site specificity.
J.Mol.Biol., 303, 2000
|
|
1FQ5
 
 | | X-ray structure of a cyclic statine inhibitor PD-129,541 bound to yeast proteinase A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(5S,9S,10S,13S)-9-hydroxy-5,10-bis(2-methylpropyl)-4,7,12,16-tetraoxo-3,6,11,17-tetraazabicyclo[17.3.1]tricosa-1(23),19,21-trien-13-yl]-3-(naphthalen-1-yl)-2-(naphthalen-1-ylmethyl)propanamide, SACCHAROPEPSIN, ... | | Authors: | Cronin, N.B, Badasso, M.O, Tickle, I.J, Dreyer, T, Hoover, D.J, Rosati, R.L, Humblet, C.C, Lunney, E.A, Cooper, J.B. | | Deposit date: | 2000-09-03 | | Release date: | 2000-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of five renin inhibitors bound to saccharopepsin: exploration of active-site specificity.
J.Mol.Biol., 303, 2000
|
|
4MLQ
 
 | | Crystal structure of Bacillus megaterium porphobilinogen deaminase | | Descriptor: | 3-[(5S)-5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]propanoic acid, 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, ACETIC ACID, ... | | Authors: | Azim, N, Deery, E, Warren, M.J, Erskine, P, Cooper, J.B, Coker, A, Wood, S.P, Akhtar, M. | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural evidence for the partially oxidized dipyrromethene and dipyrromethanone forms of the cofactor of porphobilinogen deaminase: structures of the Bacillus megaterium enzyme at near-atomic resolution.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1EPM
 
 | | A STRUCTURAL COMPARISON OF 21 INHIBITOR COMPLEXES OF THE ASPARTIC PROTEINASE FROM ENDOTHIA PARASITICA | | Descriptor: | ENDOTHIAPEPSIN, PS2, THR-PHE-GLN-ALA-PSA-LEU-ARG-GLU, ... | | Authors: | Crawford, M, Cooper, J.B, Strop, P, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
1FQ4
 
 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN HYDROXYETHYLENE INHIBITOR CP-108,420 AND YEAST ASPARTIC PROTEINASE A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(2R)-1-{[(2S,3R,5R)-1-cyclohexyl-3-hydroxy-5-{[2-(morpholin-4-yl)ethyl]carbamoyl}oct-7-yn-2-yl]amino}-3-(methylsulfa nyl)-1-oxopropan-2-yl]-1H-benzimidazole-2-carboxamide, SACCHAROPEPSIN, ... | | Authors: | Cronin, N.B, Badasso, M.O, Tickle, I.J, Dreyer, T, Hoover, D.J, Rosati, R.L, Humblet, C.C, Lunney, E.A, Cooper, J.B. | | Deposit date: | 2000-09-03 | | Release date: | 2000-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray structures of five renin inhibitors bound to saccharopepsin: exploration of active-site specificity.
J.Mol.Biol., 303, 2000
|
|
1EPL
 
 | | A STRUCTURAL COMPARISON OF 21 INHIBITOR COMPLEXES OF THE ASPARTIC PROTEINASE FROM ENDOTHIA PARASITICA | | Descriptor: | ENDOTHIAPEPSIN, PS1, PRO-LEU-GLU-PSA-ARG-LEU | | Authors: | Al-Karadaghi, S, Cooper, J.B, Strop, P, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
1EPR
 
 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-135,040 | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide | | Authors: | Badasso, M, Crawford, M, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
1GN2
 
 | | S123C mutant of the iron-superoxide dismutase from Mycobacterium tuberculosis. | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE | | Authors: | Bunting, K.A, Cooper, J.B, Tickle, I.J, Young, D.B. | | Deposit date: | 2001-10-02 | | Release date: | 2001-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Engineering of an Intersubunit Disulfide Bridge in the Iron-Superoxide Dismutase of Mycobacterium Tuberculosis.
Arch.Biochem.Biophys., 397, 2002
|
|
1EB3
 
 | | YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE 4,7-DIOXOSEBACIC ACID COMPLEX | | Descriptor: | 4,7-DIOXOSEBACIC ACID, 5-AMINOLAEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Coates, L, Newbold, R, Brindley, A.A, Stauffer, F, Wood, S.P, Warren, M.J, Cooper, J.B, Shoolingin-Jordan, P.M, Neier, R. | | Deposit date: | 2001-07-18 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Two Diacid Inhibitors
FEBS Lett., 503, 2001
|
|
1E51
 
 | | Crystal structure of native human erythrocyte 5-aminolaevulinic acid dehydratase | | Descriptor: | 3-[5-(AMINOMETHYL)-4-(CARBOXYMETHYL)-1H-PYRROL-3-YL]PROPANOIC ACID, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, SULFATE ION, ... | | Authors: | Mills-Davies, N.L, Thompson, D, Cooper, J.B, Shoolingin-Jordan, P.M. | | Deposit date: | 2000-07-13 | | Release date: | 2001-07-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | The Crystal Structure of Human Ala-Dehydratase
To be Published
|
|
1GKT
 
 | | Neutron Laue diffraction structure of endothiapepsin complexed with transition state analogue inhibitor H261 | | Descriptor: | ENDOTHIAPEPSIN, INHIBITOR, H261 | | Authors: | Coates, L, Erskine, P.T, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2001-08-20 | | Release date: | 2001-11-20 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | A Neutron Laue Diffraction Study of Endothiapepsin: Implications for the Aspartic Proteinase Mechanism
Biochemistry, 40, 2001
|
|
1GJP
 
 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH 4-OXOSEBACIC ACID | | Descriptor: | 4-OXODECANEDIOIC ACID, 5-AMINOLAEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Coates, L, Newbold, R, Brindley, A.A, Wood, S.P, Warren, M.J, Cooper, J.B, Shoolingin-Jordan, P.M, Neier, R. | | Deposit date: | 2001-08-01 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Two Diacid Inhibitors
FEBS Lett., 503, 2001
|
|
1GN4
 
 | | H145E mutant of Mycobacterium tuberculosis iron-superoxide dismutase. | | Descriptor: | MANGANESE (II) ION, SUPEROXIDE DISMUTASE | | Authors: | Bunting, K.A, Cooper, J.B, Badasso, M.O, Tickle, I.J, Newton, M, Wood, S.P, Zhang, Y, Young, D.B. | | Deposit date: | 2001-10-02 | | Release date: | 2001-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineering a Change in the Metal-Ion Specificity of the Iron-Depedent Superoxide Dismutase from Mycobacterium Tuberculosis. X-Ray Structure Analysis of Site-Directed Mutants
Eur.J.Biochem., 251, 1998
|
|
1GN3
 
 | | H145Q mutant of Mycobacterium tuberculosis iron-superoxide dismutase. | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE | | Authors: | Bunting, K.A, Cooper, J.B, Badasso, M.O, Tickle, I.J, Newton, M, Wood, S.P, Zhang, Y, Young, D.B. | | Deposit date: | 2001-10-02 | | Release date: | 2001-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Engineering a Change in the Metal-Ion Specificity of the Iron-Depedent Superoxide Dismutase from Mycobacterium Tuberculosis. X-Ray Structure Analysis of Site-Directed Mutants.
Eur.J.Biochem., 251, 1998
|
|
