4M0S
 
 | |
4XKM
 
 | | Crystal structure of Xylose Isomerase from an human intestinal tract microbe Bacteroides thetaiotaomicron | | Descriptor: | MANGANESE (II) ION, Xylose isomerase | | Authors: | Han, B.G, Bong, S.M, Cho, J.W, Lee, B.I. | | Deposit date: | 2015-01-12 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a class 2 D-xylose isomerase from the human intestinal tract microbe Bacteroides thetaiotaomicron
Biodesign, 3, 2015
|
|
4JX7
 
 | | Crystal structure of Pim1 kinase in complex with inhibitor 2-[(trans-4-aminocyclohexyl)amino]-4-{[3-(trifluoromethyl)phenyl]amino}pyrido[4,3-d]pyrimidin-5(6H)-one | | Descriptor: | 2-[(trans-4-aminocyclohexyl)amino]-4-{[3-(trifluoromethyl)phenyl]amino}pyrido[4,3-d]pyrimidin-5(6H)-one, PIM1 consensus peptide, Serine/threonine-protein kinase pim-1 | | Authors: | Lee, S.J, Han, B.G, Cho, J.W, Choi, J.S, Lee, J.K, Song, H.J, Koh, J.S, Lee, B.I. | | Deposit date: | 2013-03-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of pim1 kinase in complex with a pyrido[4,3-d]pyrimidine derivative suggests a unique binding mode.
Plos One, 8, 2013
|
|
4JX3
 
 | | Crystal structure of Pim1 kinase | | Descriptor: | Serine/threonine-protein kinase pim-1 | | Authors: | Lee, S.J, Han, B.G, Cho, J.W, Choi, J.S, Lee, J.K, Song, H.J, Koh, J.S, Lee, B.I. | | Deposit date: | 2013-03-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of pim1 kinase in complex with a pyrido[4,3-d]pyrimidine derivative suggests a unique binding mode.
Plos One, 8, 2013
|
|
3U0R
 
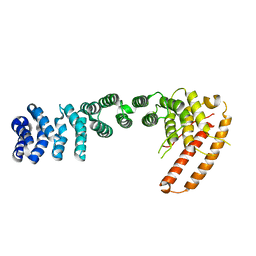 | | Helical repeat structure of apoptosis inhibitor 5 reveals protein-protein interaction modules | | Descriptor: | Apoptosis inhibitor 5 | | Authors: | Han, B.G, Kim, K.H, Jeong, K.C, Cho, J.W, Noh, K.H, Kim, T.W, Yoon, H.J, Suh, S.W, Lee, S.H, Lee, B.I. | | Deposit date: | 2011-09-29 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Helical repeat structure of apoptosis inhibitor 5 reveals protein-protein interaction modules.
J.Biol.Chem., 287, 2012
|
|
3TIO
 
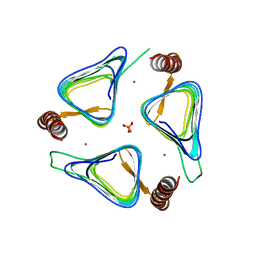 | | Crystal structures of yrdA from Escherichia coli, a homologous protein of gamma-class carbonic anhydrase, show possible allosteric conformations | | Descriptor: | PHOSPHATE ION, Protein YrdA, ZINC ION | | Authors: | Park, H.M, Choi, J.W, Lee, J.E, Jung, C.H, Kim, B.Y, Kim, J.S. | | Deposit date: | 2011-08-21 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structures of the gamma-class carbonic anhydrase homologue YrdA suggest a possible allosteric switch
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3BM1
 
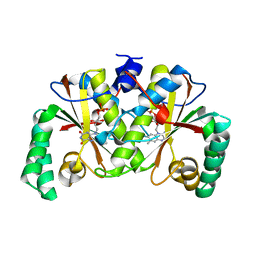 | |
3BM2
 
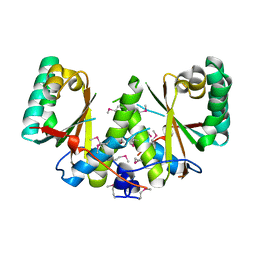 | |
4L62
 
 | |
3LY6
 
 | |
5H60
 
 | | Structure of Transferase mutant-C23S,C199S | | Descriptor: | MANGANESE (II) ION, Transferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Park, J.B, Yoo, Y, Kim, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-12-20 | | Last modified: | 2018-10-31 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
5H61
 
 | | Structure of Transferase mutant-C23S,C199S | | Descriptor: | Transferase | | Authors: | Park, J.B, Yoo, Y, Kim, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
5H5Y
 
 | | Structure of Transferase mutant-C23S,C199S | | Descriptor: | Non-LEE encoded effector protein NleB | | Authors: | Park, J.B, Yoo, Y, Kim, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
5H63
 
 | | Structure of Transferase mutant-C23S,C199S | | Descriptor: | MANGANESE (II) ION, Transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Park, J.B, Yoo, Y, Kim, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
5H62
 
 | | Structure of Transferase mutant-C23S,C199S | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, Transferase, ... | | Authors: | Park, J.B, Yoo, Y, Kim, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
5C16
 
 | | Myotubularin-related proetin 1 | | Descriptor: | Myotubularin-related protein 1, PHOSPHATE ION | | Authors: | Lee, B.I, Bong, S.M. | | Deposit date: | 2015-06-13 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structure of Human Myotubularin-Related Protein 1 Provides Insight into the Structural Basis of Substrate Specificity
Plos One, 11, 2016
|
|
3V6A
 
 | |
3V68
 
 | |
7THM
 
 | | SARS-CoV-2 nsp12/7/8 complex with a native N-terminus nsp9 | | Descriptor: | MANGANESE (II) ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Osinski, A, Tagliabracci, V.S, Chen, Z, Li, Y. | | Deposit date: | 2022-01-11 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The mechanism of RNA capping by SARS-CoV-2.
Nature, 609, 2022
|
|
3TIS
 
 | | Crystal structures of yrdA from Escherichia coli, a homologous protein of gamma-class carbonic anhydrases, show possible allosteric conformations | | Descriptor: | Protein YrdA, ZINC ION | | Authors: | Park, H.M, Chio, J.W, Lee, J.E, Jung, J.H, Kim, B.Y, Kim, J.S. | | Deposit date: | 2011-08-21 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the gamma-class carbonic anhydrase homologue YrdA suggest a possible allosteric switch
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3WUT
 
 | | Structure basis of inactivating cell abscission | | Descriptor: | Centrosomal protein of 55 kDa, GLYCEROL, Inactive serine/threonine-protein kinase TEX14 | | Authors: | Kim, H.J, Matsuura, A, Lee, H.H. | | Deposit date: | 2014-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural and biochemical insights into the role of testis-expressed gene 14 (TEX14) in forming the stable intercellular bridges of germ cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3WUV
 
 | | Structure basis of inactivating cell abscission with chimera peptide 2 | | Descriptor: | Centrosomal protein of 55 kDa, peptide from Programmed cell death 6-interacting protein | | Authors: | Kim, H.J, Matsuura, A, Lee, H.H. | | Deposit date: | 2014-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and biochemical insights into the role of testis-expressed gene 14 (TEX14) in forming the stable intercellular bridges of germ cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3WUU
 
 | |
3H1T
 
 | |
4MCV
 
 | | Star 12 bound to analog-sensitive Src kinase | | Descriptor: | (7S)-12-(4-aminobutyl)-7-(2-methylpropyl)-6,7,12,13-tetrahydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazol-5-one, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Lopez, M.S, Shokat, K.M. | | Deposit date: | 2013-08-21 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Staurosporine-derived inhibitors broaden the scope of analog-sensitive kinase technology.
J.Am.Chem.Soc., 135, 2013
|
|
