1EA2
 
 | | Pseudoreversion of the Catalytic Activity of Y14F by the Additional Tyrosine-to-Phenylalanine Substitution(s) in the Hydrogen Bond Network of Delta-5-3-Ketosteroid Isomerase from Pheudomonas putida Biotype B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Choi, G, Ha, N.-C, Kim, M.-S, Hong, B.-H, Choi, K.Y. | | Deposit date: | 2000-11-03 | | Release date: | 2001-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pseudoreversion of the Catalytic Activity of Y14F by the Additional Substitution(S) of Tyrosine with Phenylalanine in the Hydrogen Bond Network of Delta (5)-3-Ketosteroid Isomerase from Pseudomonas Putida Biotype B
Biochemistry, 40, 2001
|
|
6FJM
 
 | | tubulin-Disorazole Z complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Disorazole Z, ... | | Authors: | Menchon, G, Prota, A.E, Lucena Agell, D, Bucher, P, Jansen, R, Irschik, H, Mueller, R, Paterson, I, Diaz, J.F, Altmann, K.-H, Steinmetz, M.O. | | Deposit date: | 2018-01-22 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A fluorescence anisotropy assay to discover and characterize ligands targeting the maytansine site of tubulin.
Nat Commun, 9, 2018
|
|
6FII
 
 | | Tubulin-Spongistatin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Menchon, G, Prota, A.E, Lucena Angell, D, Bucher, P, Mueller, R, Paterson, I, Diaz, J.F, Altmann, K.-H, Steinmetz, M.O. | | Deposit date: | 2018-01-18 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | A fluorescence anisotropy assay to discover and characterize ligands targeting the maytansine site of tubulin.
Nat Commun, 9, 2018
|
|
6FJF
 
 | | Tubulin-FcMaytansine complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Menchon, G, Prota, A.E, Lucena Angell, D, Bucher, P, Mueller, R, Paterson, I, Diaz, J.F, Altmann, K.-H, Steinmetz, M.O. | | Deposit date: | 2018-01-22 | | Release date: | 2018-05-30 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | A fluorescence anisotropy assay to discover and characterize ligands targeting the maytansine site of tubulin.
Nat Commun, 9, 2018
|
|
8BX1
 
 | |
8BX2
 
 | |
1CQS
 
 | | CRYSTAL STRUCTURE OF D103E MUTANT WITH EQUILENINEOF KSI IN PSEUDOMONAS PUTIDA | | Descriptor: | EQUILENIN, PROTEIN : KETOSTEROID ISOMERASE | | Authors: | Choi, G, Ha, N.C, Kim, S.W, Kim, D.H, Park, S, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-08-11 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Asp-99 donates a hydrogen bond not to Tyr-14 but to the steroid directly
in the catalytic mechanism of Delta 5-3-ketosteroid isomerase from
Pseudomonas putida biotype B
Biochemistry, 39, 2000
|
|
1LN6
 
 | | STRUCTURE OF BOVINE RHODOPSIN (Metarhodopsin II) | | Descriptor: | RETINAL, RHODOPSIN | | Authors: | Choi, G, Landin, J, Galan, J.F, Birge, R.R, Albert, A.D, Yeagle, P.L. | | Deposit date: | 2002-05-03 | | Release date: | 2002-07-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural studies of metarhodopsin II, the activated form of the G-protein coupled receptor, rhodopsin.
Biochemistry, 41, 2002
|
|
5OV7
 
 | | tubulin - rigosertib complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Menchon, G, Prota, A.E, Steinmetz, M, Jost, M. | | Deposit date: | 2017-08-28 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Combined CRISPRi/a-Based Chemical Genetic Screens Reveal that Rigosertib Is a Microtubule-Destabilizing Agent.
Mol. Cell, 68, 2017
|
|
8H1A
 
 | |
8H27
 
 | |
8H0S
 
 | |
8H0T
 
 | |
8H1B
 
 | | Crystal structure of MnmM from S. aureus complexed with SAM and tRNA anti-codon stem loop (ASL) (1.55 A) | | Descriptor: | RNA (5'-R(*AP*CP*GP*GP*AP*CP*UP*UP*UP*GP*AP*CP*UP*CP*CP*GP*U)-3'), S-ADENOSYLMETHIONINE, SODIUM ION, ... | | Authors: | Kim, J, Cho, G, Lee, J. | | Deposit date: | 2022-10-01 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of a novel 5-aminomethyl-2-thiouridine methyltransferase in tRNA modification.
Nucleic Acids Res., 51, 2023
|
|
7CNX
 
 | | Crystal structure of Apo PSD from E. coli (2.63 A) | | Descriptor: | Phosphatidylserine decarboxylase alpha chain, Phosphatidylserine decarboxylase beta chain | | Authors: | Kim, J, Cho, G. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural insights into phosphatidylethanolamine formation in bacterial membrane biogenesis.
Sci Rep, 11, 2021
|
|
7CNY
 
 | | Crystal structure of 8PE bound PSD from E. coli (2.12 A) | | Descriptor: | 1,2-Dioctanoyl-SN-Glycero-3-Phosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, Phosphatidylserine decarboxylase alpha chain, ... | | Authors: | Kim, J, Cho, G. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural insights into phosphatidylethanolamine formation in bacterial membrane biogenesis.
Sci Rep, 11, 2021
|
|
7CNZ
 
 | | Crystal structure of 10PE bound PSD from E. coli (2.70 A) | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, PHOSPHATE ION, Phosphatidylserine decarboxylase alpha chain, ... | | Authors: | Kim, J, Cho, G. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into phosphatidylethanolamine formation in bacterial membrane biogenesis.
Sci Rep, 11, 2021
|
|
7CNW
 
 | | Crystal structure of Apo PSD from E. coli (1.90 A) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Phosphatidylserine decarboxylase alpha chain, Phosphatidylserine decarboxylase beta chain, ... | | Authors: | Kim, J, Cho, G. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into phosphatidylethanolamine formation in bacterial membrane biogenesis.
Sci Rep, 11, 2021
|
|
2CF4
 
 | | Pyrococcus horikoshii TET1 peptidase can assemble into a tetrahedron or a large octahedral shell | | Descriptor: | COBALT (II) ION, PROTEIN PH0519 | | Authors: | Vellieux, F.M.D, Schoehn, G, Dura, M.A, Roussel, A, Franzetti, B. | | Deposit date: | 2006-02-15 | | Release date: | 2006-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | An Archaeal Peptidase Assembles Into Two Different Quaternary Structures: A Tetrahedron and a Giant Octahedron.
J.Biol.Chem., 281, 2006
|
|
7Q5R
 
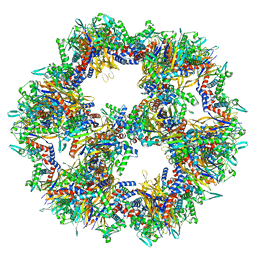 | | Protein community member pyruvate dehydrogenase complex E2 core from C. thermophilum | | Descriptor: | Acetyltransferase component of pyruvate dehydrogenase complex | | Authors: | Chojnowski, G, Skalidis, I, Kyrilis, F.L, Tueting, C, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM and artificial intelligence visualize endogenous protein community members.
Structure, 30, 2022
|
|
7Q5Q
 
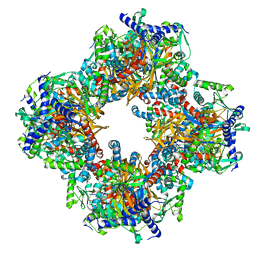 | | Protein community member oxoglutarate dehydrogenase complex E2 core from C. thermophilum | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase | | Authors: | Chojnowski, G, Skalidis, I, Kyrilis, F.L, Tueting, C, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | Cryo-EM and artificial intelligence visualize endogenous protein community members.
Structure, 30, 2022
|
|
7Q5S
 
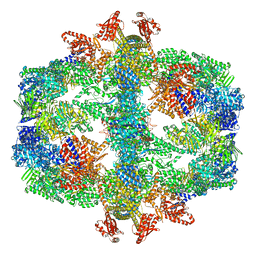 | | Protein community member fatty acid synthase complex from C. thermophilum | | Descriptor: | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase, 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Chojnowski, G, Skalidis, I, Kyrilis, F.L, Tueting, C, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | Cryo-EM and artificial intelligence visualize endogenous protein community members.
Structure, 30, 2022
|
|
7B9F
 
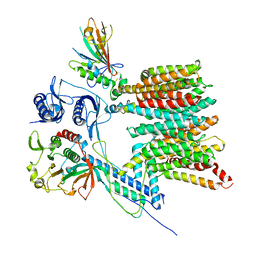 | | Structure of the mycobacterial ESX-5 Type VII Secretion System hexameric pore complex | | Descriptor: | EccB5, EccC5, EccD5, ... | | Authors: | Chojnowski, G, Ritter, C, Beckham, K.S.H, Mullapudi, E, Rettel, M, Savitski, M.M, Mortensen, S.A, Ziemianowicz, D, Kosinski, J, Wilmanns, M. | | Deposit date: | 2020-12-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the mycobacterial ESX-5 type VII secretion system pore complex.
Sci Adv, 7, 2021
|
|
7B9S
 
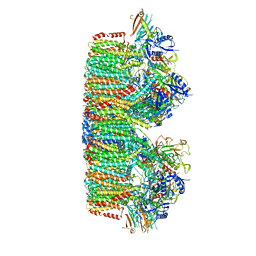 | | Structure of the mycobacterial ESX-5 Type VII Secretion System hexameric pore complex | | Descriptor: | EccB5, EccC5, EccD5, ... | | Authors: | Chojnowski, G, Ritter, C, Beckham, K.S.H, Mullapudi, E, Rettel, M, Savitski, M.M, Mortensen, S.A, Ziemianowicz, D, Kosinski, J, Wilmanns, M. | | Deposit date: | 2020-12-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the mycobacterial ESX-5 type VII secretion system pore complex.
Sci Adv, 7, 2021
|
|
5OSK
 
 | | Tubulin-7j complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-(2,5-Dimethoxybenzyl)-7-sulfamoyloxy-6-methoxy-3,4-dihydroquinazolin-2(1H)-one, CALCIUM ION, ... | | Authors: | Menchon, G, Prota, A.E, Steinmetz, M.O, Potter, B.V.L. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Quinazolinone-Based Anticancer Agents: Synthesis, Antiproliferative SAR, Antitubulin Activity, and Tubulin Co-crystal Structure.
J. Med. Chem., 61, 2018
|
|
