4MY4
 
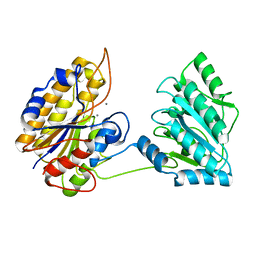 | | Crystal structure of phosphoglycerate mutase from Staphylococcus aureus. | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, MANGANESE (II) ION | | Authors: | Roychowdhury, A, Kundu, A, Gujar, A, Bose, M, Das, A.K. | | Deposit date: | 2013-09-27 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complete catalytic cycle of cofactor-independent phosphoglycerate mutase involves a spring-loaded mechanism
Febs J., 282, 2015
|
|
4W5C
 
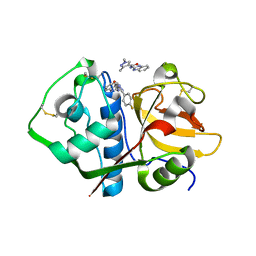 | | Crystal structure analysis of cruzain with three Fragments: 1 (N-(1H-benzimidazol-2-yl)-1,3-dimethyl-pyrazole-4-carboxamide), 6 (2-amino-4,6-difluorobenzothiazole) and 9 (N-(1H-benzimidazol-2-yl)-3-(4-fluorophenyl)-1H-pyrazole-4-carboxamide). | | Descriptor: | 4,6-difluoro-1,3-benzothiazol-2-amine, Cruzipain, N-(1H-benzimidazol-2-yl)-1,3-dimethyl-1H-pyrazole-4-carboxamide, ... | | Authors: | Tochowicz, A, McKerrow, J.H, Craik, C.S. | | Deposit date: | 2014-08-17 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Applying Fragments Based- Drug Design to identify multiple binding modes on cysteine protease.
To Be Published
|
|
7M3Q
 
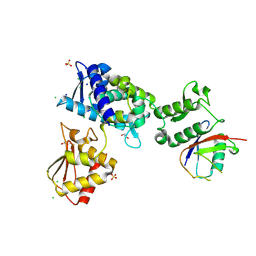 | | Structure of the Smurf2 HECT Domain with a High Affinity Ubiquitin Variant (UbV) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, ... | | Authors: | Chowdhury, A, Singer, A.U, Ogunjimi, A.A, Teyra, J, Zhang, W, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-03-18 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Smurf2 HECT Domain with a High Affinity Ubiquitin Variant (UbV)
To be published
|
|
4TRO
 
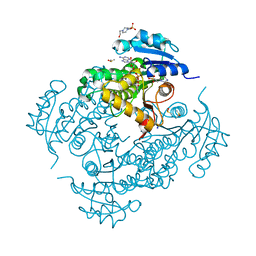 | | Structure of the enoyl-ACP reductase of Mycobacterium tuberculosis InhA, inhibited with the active metabolite of isoniazid | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Chollet, A, Julien, S, Mourey, L, Maveyraud, L. | | Deposit date: | 2014-06-17 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the enoyl-ACP reductase of Mycobacterium tuberculosis (InhA) in the apo-form and in complex with the active metabolite of isoniazid pre-formed by a biomimetic approach.
J.Struct.Biol., 190, 2015
|
|
4TRM
 
 | | Structure of the apo form of InhA from Mycobacterium tuberculosis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Chollet, A, Julien, S, Mourey, L, Maveyraud, L. | | Deposit date: | 2014-06-17 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the enoyl-ACP reductase of Mycobacterium tuberculosis (InhA) in the apo-form and in complex with the active metabolite of isoniazid pre-formed by a biomimetic approach.
J.Struct.Biol., 190, 2015
|
|
4TRN
 
 | | STRUCTURE OF INHA FROM MYCOBACTERIUM TUBERCULOSIS COMPLEXED TO NADH | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, INHA, ... | | Authors: | Chollet, A, Julien, S, Mourey, L, Maveyraud, L. | | Deposit date: | 2014-06-17 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the enoyl-ACP reductase of Mycobacterium tuberculosis (InhA) in the apo-form and in complex with the active metabolite of isoniazid pre-formed by a biomimetic approach.
J.Struct.Biol., 190, 2015
|
|
4W5B
 
 | |
1BII
 
 | | THE CRYSTAL STRUCTURE OF H-2DD MHC CLASS I IN COMPLEX WITH THE HIV-1 DERIVED PEPTIDE P18-110 | | Descriptor: | BETA-2 MICROGLOBULIN, DECAMERIC PEPTIDE, MHC CLASS I H-2DD | | Authors: | Achour, A, Persson, K, Harris, R.A, Sundback, J, Sentman, C.L, Lindqvist, Y, Schneider, G, Karre, K. | | Deposit date: | 1998-06-11 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of H-2Dd MHC class I complexed with the HIV-1-derived peptide P18-I10 at 2.4 A resolution: implications for T cell and NK cell recognition.
Immunity, 9, 1998
|
|
8DXB
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 5-fluoroindole-2-carboxylic acid at the NNRTI Adjacent site | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, 5-fluoroindole-2-carboxylic acid, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DXH
 
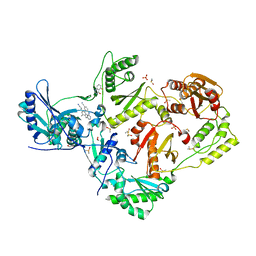 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 5-fluoroquinazolin-4-ol at W266 site | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, 5-fluoranyl-3,4-dihydroquinazolin-4-ol, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DX8
 
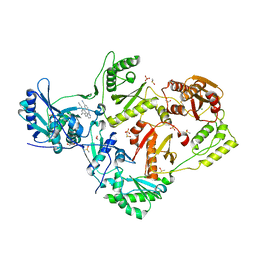 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 2-chloro-6-fluorophenethylamine at the 415 site | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-chloro-6-fluorophenyl)ethan-1-amine, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DXM
 
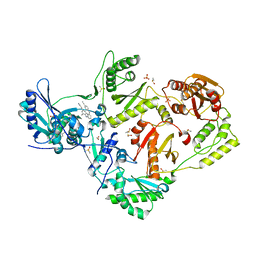 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 4-bromophenol at the Knuckles site | | Descriptor: | 1,2-ETHANEDIOL, 4-BROMOPHENOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DXI
 
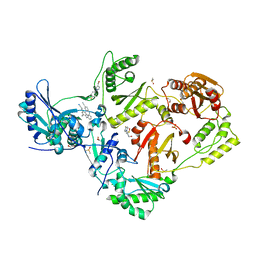 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment [1-(4-fluorophenyl)-5-methyl-1H-pyrazol-4-yl]methanol at multiple sites | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DXK
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 4-bromopyrazole at multiple sites | | Descriptor: | 1,2-ETHANEDIOL, 4-bromo-1H-pyrazole, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DXG
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 5-(trifluoromethyl)pyridin-2-ol at W24 site | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, 5-(trifluoromethyl)pyridin-2-one, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DXL
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 4-iodopyrazole at multiple sites | | Descriptor: | 1,2-ETHANEDIOL, 4-IODOPYRAZOLE, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DX2
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 4-amino-3-bromopyridine at multiple sites | | Descriptor: | 1,2-ETHANEDIOL, 3-bromopyridin-4-amine, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DX3
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 3-bromobenzylamine in the thumb subdomain | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-bromophenyl)methanamine, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DXE
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 2-amino-6-fluorobenzonitrile at the NNRTI adjacent site | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-6-fluorobenzonitrile, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DXJ
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 1-N-methyl-4-(trifluoromethyl)benzene-1,2-diamine at the NNRTI adjacent site | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
6H57
 
 | | Crystal structure of S. cerevisiae DEAH-box RNA helicase Dhr1, essential for small ribosomal subunit biogenesis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Roychowdhury, A, Graille, M. | | Deposit date: | 2018-07-24 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The DEAH-box RNA helicase Dhr1 contains a remarkable carboxyl terminal domain essential for small ribosomal subunit biogenesis.
Nucleic Acids Res., 47, 2019
|
|
2F74
 
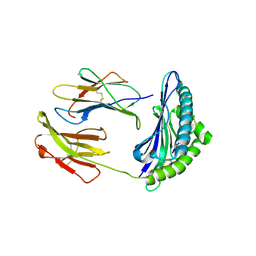 | | Murine MHC class I H-2Db in complex with human b2-microglobulin and LCMV-derived immunodminant peptide gp33 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Achour, A, Michaelsson, J, Harris, R.A, Ljunggren, H.G, Karre, K, Schneider, G, Sandalova, T. | | Deposit date: | 2005-11-30 | | Release date: | 2006-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of the Differential Stability and Receptor Specificity of H-2D(b) in Complex with Murine versus Human beta(2)-Microglobulin.
J.Mol.Biol., 356, 2006
|
|
6I8C
 
 | | Crystal structure of the murine beta-2-microglobulin. | | Descriptor: | Beta-2-microglobulin, TRIETHYLENE GLYCOL | | Authors: | Achour, A, Sandalova, T, Ricagno, S, Sun, R. | | Deposit date: | 2018-11-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biochemical and biophysical comparison of human and mouse beta-2 microglobulin reveals the molecular determinants of low amyloid propensity.
Febs J., 287, 2020
|
|
4ISK
 
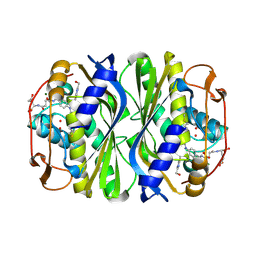 | | Crystal structure of E.coli thymidylate synthase with dUMP and the BGC 945 inhibitor | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2'-deoxy-5'-uridylic acid, MAGNESIUM ION, ... | | Authors: | Tochowicz, A, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2013-01-16 | | Release date: | 2013-12-25 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Development and Binding Mode Assessment of N-[4-[2-Propyn-1-yl[(6S)-4,6,7,8-tetrahydro-2-(hydroxymethyl)-4-oxo-3H-cyclopenta[g]quinazolin-6-yl]amino]benzoyl]-l-gamma-glutamyl-d-glutamic Acid (BGC 945), a Novel Thymidylate Synthase Inhibitor That Targets Tumor Cells.
J.Med.Chem., 56, 2013
|
|
7ZIT
 
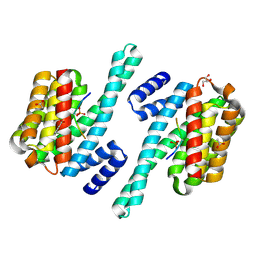 | | 14-3-3 in complex with SARS-COV2 N phospho-peptide | | Descriptor: | 14-3-3 protein zeta/delta, ACETATE ION, BENZOIC ACID, ... | | Authors: | Eisenreichova, A, Boura, E. | | Deposit date: | 2022-04-08 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural basis for SARS-CoV-2 nucleocapsid (N) protein recognition by 14-3-3 proteins.
J.Struct.Biol., 214, 2022
|
|
