8H93
 
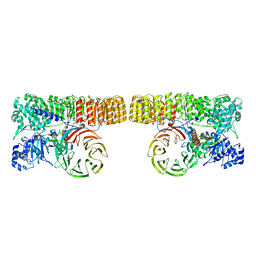 | | Structure of dimeric mouse SCMC core complex | | Descriptor: | NACHT, LRR and PYD domains-containing protein 5, Oocyte-expressed protein homolog, ... | | Authors: | Chi, P, Ou, G, Han, Z, Li, J, Deng, D. | | Deposit date: | 2022-10-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis of the subcortical maternal complex and its implications in reproductive disorders.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H94
 
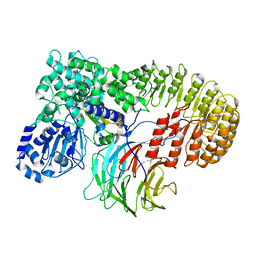 | | Structure of mouse SCMC bound with KH domain of FILIA | | Descriptor: | NACHT, LRR and PYD domains-containing protein 5, Oocyte-expressed protein homolog, ... | | Authors: | Chi, P, Ou, G, Han, Z, Li, J, Deng, D. | | Deposit date: | 2022-10-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of the subcortical maternal complex and its implications in reproductive disorders.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H95
 
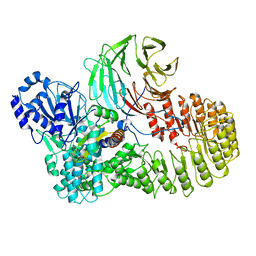 | | Structure of mouse SCMC bound with full-length FILIA | | Descriptor: | NACHT, LRR and PYD domains-containing protein 5, Oocyte-expressed protein homolog, ... | | Authors: | Chi, P, Ou, G, Han, Z, Li, L, Deng, D. | | Deposit date: | 2022-10-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis of the subcortical maternal complex and its implications in reproductive disorders.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H96
 
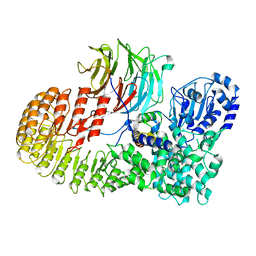 | | Structure of mouse SCMC core complex | | Descriptor: | NACHT, LRR and PYD domains-containing protein 5, Oocyte-expressed protein homolog, ... | | Authors: | Chi, P, Ou, G, Li, J, Han, Z, Deng, D. | | Deposit date: | 2022-10-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structural basis of the subcortical maternal complex and its implications in reproductive disorders.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H8F
 
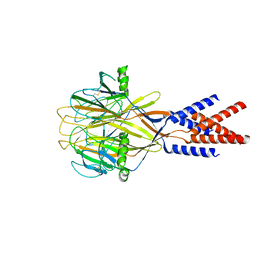 | | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR (resting state) | | Descriptor: | Proton-activated chloride channel | | Authors: | Chi, P, Wang, X, Li, J, Li, K, Zhang, Y, Geng, J, Wu, J, Deng, D. | | Deposit date: | 2022-10-22 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR (resting state)
To Be Published
|
|
8H8D
 
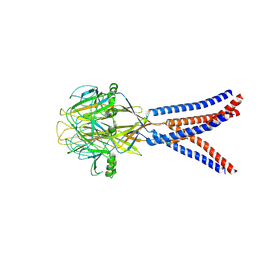 | | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR trimer bound with tRNA (intermediate state) | | Descriptor: | Proton-activated chloride channel | | Authors: | Chi, P, Wang, X, Li, J, Li, K, Zhang, Y, Geng, J, Wu, J, Deng, D. | | Deposit date: | 2022-10-22 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR trimer bound with tRNA (intermediate state)
To Be Published
|
|
8H8E
 
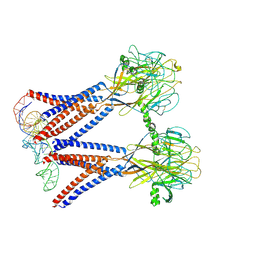 | | Structure of the dimeric Xenopus tropical acid-sensitive outwardly rectifying channel ASOR trimer bound with tRNA (closed state) | | Descriptor: | Proton-activated chloride channel, tRNA (75-MER)of Spodoptera frugiperda | | Authors: | Chi, P, Wang, X, Li, J, Li, K, Zhang, Y, Geng, J, Wu, J, Deng, D. | | Deposit date: | 2022-10-22 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structure of the dimeric Xenopus tropical acid-sensitive outwardly rectifying channel ASOR trimer bound with tRNA (closed state)
To Be Published
|
|
7FIB
 
 | |
8CP3
 
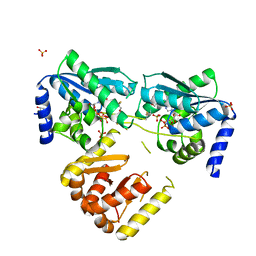 | | Apo-LarE in complex with AMP-PNP | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, NAD_synthase domain-containing protein, ... | | Authors: | Zecchin, P, Pecqueur, L, Golinelli-Pimpaneau, B. | | Deposit date: | 2023-03-01 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-based insights into the mechanism of [4Fe-4S]-dependent sulfur insertase LarE.
Protein Sci., 33, 2024
|
|
8CP4
 
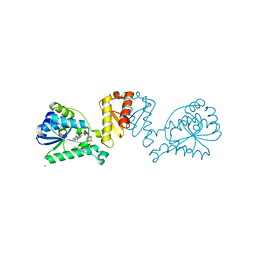 | | [4Fe-4S] cluster containing LarE in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Zecchin, P, Pecqueur, L, Golinelli-Pimpaneau, B. | | Deposit date: | 2023-03-01 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure-based insights into the mechanism of [4Fe-4S]-dependent sulfur insertase LarE.
Protein Sci., 33, 2024
|
|
4OYT
 
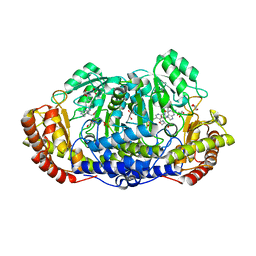 | | Crystal structure of ternary complex of Plasmodium vivax SHMT with D-serine and folinic acid | | Descriptor: | (2R)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-oxidanyl-propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U. | | Deposit date: | 2014-02-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Plasmodium vivax serine hydroxymethyltransferase: implications for ligand-binding specificity and functional control.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PFN
 
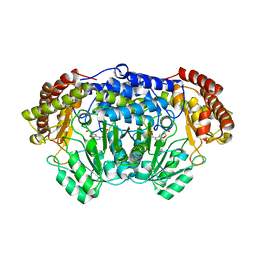 | | Crystal structure of Plasmodium vivax SHMT with L-serine Schiff base | | Descriptor: | CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, SERINE, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U. | | Deposit date: | 2014-04-30 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Plasmodium vivax serine hydroxymethyltransferase: implications for ligand-binding specificity and functional control.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3IRM
 
 | | Trypanosoma cruzi Dihydrofolate Reductase-Thymidylate Synthase COMPLEXED WITH Cycloguanil | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, ACETATE ION, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Chitnumsub, P, Yuvaniyama, J, Yuthavong, Y. | | Deposit date: | 2009-08-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of antifolate inhibition of Trypanosoma cruzi Dihydrofolate Reductase-Thymidylate Synthase
To be Published
|
|
3IRO
 
 | |
4PFF
 
 | | Crystal structure of Plasmodium vivax SHMT with PLP Schiff base | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, putative | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U. | | Deposit date: | 2014-04-29 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Plasmodium vivax serine hydroxymethyltransferase: implications for ligand-binding specificity and functional control.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3INV
 
 | | Trypanosoma cruzi Dihydrofolate Reductase-Thymidylate Synthase COMPLEXED WITH NADPH, dUMP AND C-448 ANTIFOLATE | | Descriptor: | 1-[3-(2,3-dichlorophenoxy)propoxy]-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Chitnumsub, P, Yuvaniyama, J, Yuthavong, Y. | | Deposit date: | 2009-08-13 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural basis of antifolate inhibition of Trypanosoma cruzi Dihydrofolate Reductase-Thymidylate Synthase
To be Published
|
|
3IRN
 
 | | Trypanosoma cruzi Dihydrofolate Reductase-Thymidylate Synthase COMPLEXED WITH NADPH AND Cycloguanil | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, Bifunctional dihydrofolate reductase-thymidylate synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Chitnumsub, P, Yuvaniyama, J, Yuthavong, Y. | | Deposit date: | 2009-08-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of antifolate inhibition of Trypanosoma cruzi Dihydrofolate Reductase-Thymidylate Synthase
To be Published
|
|
2QAS
 
 | |
4O6Z
 
 | | Crystal structure of serine hydroxymethyltransferase with covalently bound PLP Schiff-base from Plasmodium falciparum | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U. | | Deposit date: | 2013-12-24 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | The structure of Plasmodium falciparum serine hydroxymethyltransferase reveals a novel redox switch that regulates its activities.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2QAZ
 
 | |
6JWW
 
 | | Crystal structure of Plasmodium falciparum HPPK-DHPS S436F/A437G/A613T triple mutant with STZ-DHP | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 4-{[(2-amino-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)methyl]amino}-N-(1,3-thiazol-2-yl)benzenesulfonamide, 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase-dihydropteroate synthase, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Yuthavong, Y. | | Deposit date: | 2019-04-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance.
Febs J., 287, 2020
|
|
6JWX
 
 | | Crystal structure of Plasmodium falciparum HPPK-DHPS wild type with SDX-DHP | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 4-[(2-azanyl-4-oxidanylidene-7,8-dihydro-3~{H}-pteridin-6-yl)methylamino]-~{N}-(5,6-dimethoxypyrimidin-4-yl)benzenesulfonamide, 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase-dihydropteroate synthase, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Yuthavong, Y. | | Deposit date: | 2019-04-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance.
Febs J., 287, 2020
|
|
6JWV
 
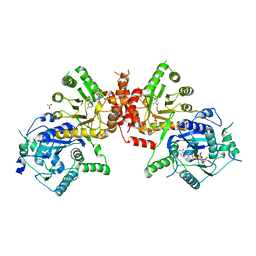 | | Crystal structure of Plasmodium falciparum HPPK-DHPS A437G with STZ-DHP | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 4-{[(2-amino-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)methyl]amino}-N-(1,3-thiazol-2-yl)benzenesulfonamide, 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase-dihydropteroate synthase, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Yuthavong, Y. | | Deposit date: | 2019-04-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance.
Febs J., 287, 2020
|
|
6JWR
 
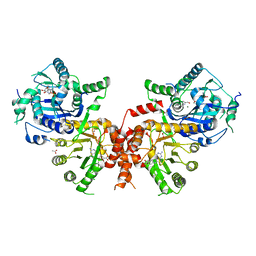 | | Crystal structure of Plasmodium falciparum HPPK-DHPS wild type with Pteroate | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase-dihydropteroate synthase, ACETATE ION, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Yuthavong, Y. | | Deposit date: | 2019-04-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance.
Febs J., 287, 2020
|
|
6JWZ
 
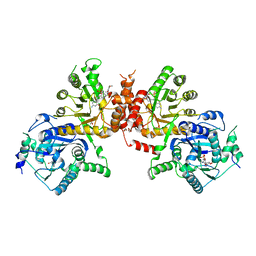 | | Crystal structure of Plasmodium falciparum HPPK-DHPS S436F/A437G/A613S triple mutant with SDX-DHP | | Descriptor: | 4-[(2-azanyl-4-oxidanylidene-7,8-dihydro-3~{H}-pteridin-6-yl)methylamino]-~{N}-(5,6-dimethoxypyrimidin-4-yl)benzenesulfonamide, 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase-dihydropteroate synthase, ACETATE ION, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Yuthavong, Y. | | Deposit date: | 2019-04-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance.
Febs J., 287, 2020
|
|
