1EZT
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF FREE RGS4 BY NMR | | Descriptor: | REGULATOR OF G-PROTEIN SIGNALING 4 | | Authors: | Moy, F.J, Chanda, P.K, Cockett, M.I, Edris, W, Jones, P.G, Mason, K, Semus, S, Powers, R. | | Deposit date: | 2000-05-11 | | Release date: | 2001-01-15 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of free RGS4 reveals an induced conformational change upon binding Galpha.
Biochemistry, 39, 2000
|
|
1EZY
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF FREE RGS4 BY NMR | | Descriptor: | REGULATOR OF G-PROTEIN SIGNALING 4 | | Authors: | Moy, F.J, Chanda, P.K, Cockett, M.I, Edris, W, Jones, P.G, Mason, K, Semus, S, Powers, R. | | Deposit date: | 2000-05-12 | | Release date: | 2001-01-15 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of free RGS4 reveals an induced conformational change upon binding Galpha.
Biochemistry, 39, 2000
|
|
1FM1
 
 | | SOLUTION STRUCTURE OF THE CATALYTIC FRAGMENT OF HUMAN COLLAGENASE-3 (MMP-13) COMPLEXED WITH A HYDROXAMIC ACID INHIBITOR | | Descriptor: | CALCIUM ION, COLLAGENASE-3, N-HYDROXY-2-[(4-METHOXY-BENZENESULFONYL)-PYRIDIN-3-YLMETHYL-AMINO]-3-METHYL-BENZAMIDE, ... | | Authors: | Moy, F.J, Chanda, P.K, Chen, J.M, Cosmi, S, Edris, W, Levin, J.I, Powers, R. | | Deposit date: | 2000-08-15 | | Release date: | 2001-08-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the catalytic fragment of human collagenase-3 (MMP-13) complexed with a hydroxamic acid inhibitor.
J.Mol.Biol., 302, 2000
|
|
1FLS
 
 | | SOLUTION STRUCTURE OF THE CATALYTIC FRAGMENT OF HUMAN COLLAGENASE-3 (MMP-13) COMPLEXED WITH A HYDROXAMIC ACID INHIBITOR | | Descriptor: | CALCIUM ION, COLLAGENASE-3, N-HYDROXY-2-[(4-METHOXY-BENZENESULFONYL)-PYRIDIN-3-YLMETHYL-AMINO]-3-METHYL-BENZAMIDE, ... | | Authors: | Moy, F.J, Chanda, P.K, Chen, J.M, Cosmi, S, Edris, W, Levin, J.I, Powers, R. | | Deposit date: | 2000-08-15 | | Release date: | 2001-08-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the catalytic fragment of human collagenase-3 (MMP-13) complexed with a hydroxamic acid inhibitor.
J.Mol.Biol., 302, 2000
|
|
3AYK
 
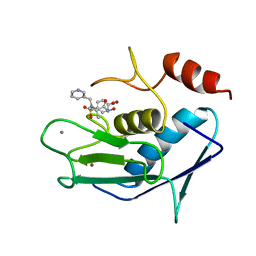 | | CATALYTIC FRAGMENT OF HUMAN FIBROBLAST COLLAGENASE COMPLEXED WITH CGS-27023A, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CALCIUM ION, N-HYDROXY-2(R)-[[(4-METHOXYPHENYL)SULFONYL](3-PICOLYL)AMINO]-3-METHYLBUTANAMIDE HYDROCHLORIDE, PROTEIN (COLLAGENASE), ... | | Authors: | Powers, R, Moy, F.J. | | Deposit date: | 1999-02-01 | | Release date: | 1999-06-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the catalytic fragment of human fibroblast collagenase complexed with a sulfonamide derivative of a hydroxamic acid compound.
Biochemistry, 38, 1999
|
|
4AYK
 
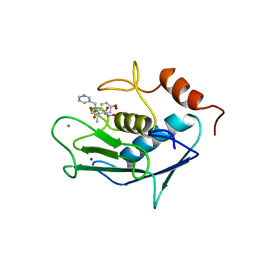 | | CATALYTIC FRAGMENT OF HUMAN FIBROBLAST COLLAGENASE COMPLEXED WITH CGS-27023A, NMR, 30 STRUCTURES | | Descriptor: | CALCIUM ION, N-HYDROXY-2(R)-[[(4-METHOXYPHENYL)SULFONYL](3-PICOLYL)AMINO]-3-METHYLBUTANAMIDE HYDROCHLORIDE, PROTEIN (COLLAGENASE), ... | | Authors: | Powers, R, Moy, F.J. | | Deposit date: | 1999-02-01 | | Release date: | 1999-06-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the catalytic fragment of human fibroblast collagenase complexed with a sulfonamide derivative of a hydroxamic acid compound.
Biochemistry, 38, 1999
|
|
1AYK
 
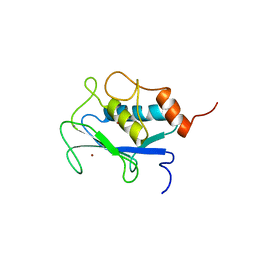 | |
2AYK
 
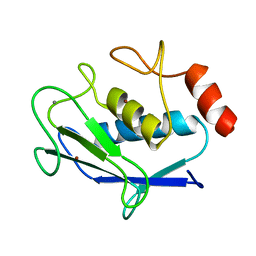 | |
5EWM
 
 | |
5EWL
 
 | |
5EWJ
 
 | |
