7YE6
 
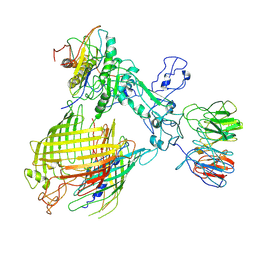 | | BAM-EspP complex structure with BamA-N427C/EspP-R1297C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
7YE4
 
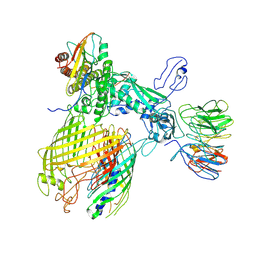 | | BAM-EspP complex structure with BamA-G431C and G781C/EspP-N1293C and A1043C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
8IEM
 
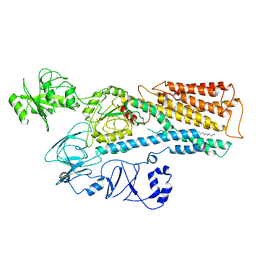 | | Cryo-EM structure of ATP13A2 in the E2P state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Polyamine-transporting ATPase 13A2, ... | | Authors: | Liu, Z.M, Mu, J.Q, Xue, C.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Conformational cycle of human polyamine transporter ATP13A2.
Nat Commun, 14, 2023
|
|
8IEN
 
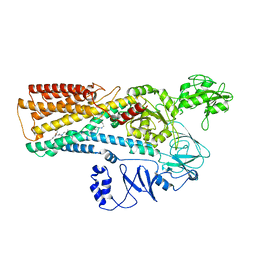 | | Cryo-EM structure of ATP13A2 in the E2-Pi state | | Descriptor: | MAGNESIUM ION, Polyamine-transporting ATPase 13A2, SPERMINE, ... | | Authors: | Liu, Z.M, Mu, J.Q, Xue, C.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Conformational cycle of human polyamine transporter ATP13A2.
Nat Commun, 14, 2023
|
|
8IES
 
 | | Cryo-EM structure of ATP13A2 in the E1P-ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Polyamine-transporting ATPase 13A2, ... | | Authors: | Liu, Z.M, Mu, J.Q, Xue, C.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Conformational cycle of human polyamine transporter ATP13A2.
Nat Commun, 14, 2023
|
|
8IEO
 
 | | Cryo-EM structure of ATP13A2 in the nominal E1P state | | Descriptor: | MAGNESIUM ION, Polyamine-transporting ATPase 13A2, SPERMINE, ... | | Authors: | Liu, Z.M, Mu, J.Q, Xue, C.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Conformational cycle of human polyamine transporter ATP13A2.
Nat Commun, 14, 2023
|
|
8IER
 
 | |
8IEL
 
 | |
8IEK
 
 | |
8BO2
 
 | | BAM-EspP complex structure with BamA-S425C/EspP-S1299C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
8BNZ
 
 | | BAM-EspP complex structure with BamA-G431C/EspP-N1293C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
6KYL
 
 | | Crystal Structure of Phosphatidic acid Transporter Ups1/Mdm35 in Complex with (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, Mitochondrial distribution and morphology protein 35, Protein UPS1, ... | | Authors: | Lu, J, Chan, K.C, Zhai, Y, Fan, J, Sun, F. | | Deposit date: | 2019-09-19 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Molecular mechanism of mitochondrial phosphatidate transfer by Ups1.
Commun Biol, 3, 2020
|
|
5JQM
 
 | | Crystal Structure of Phosphatidic acid Transporter Ups1/Mdm35 Void of Bound Phospholipid from Saccharomyces Cerevisiae at 1.5 Angstroms Resolution | | Descriptor: | Protein UPS1, mitochondrial,Mitochondrial distribution and morphology protein 35 | | Authors: | Lu, J, Chan, K.C, Zhai, Y, Fan, J, Sun, F. | | Deposit date: | 2016-05-05 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular mechanism of mitochondrial phosphatidate transfer by Ups1
Commun Biol, 2020
|
|
5JQL
 
 | | Crystal Structure of Phosphatidic acid Transporter Ups1/Mdm35 Void of Bound Phospholipid from Saccharomyces Cerevisiae at 2.9 Angstroms Resolution | | Descriptor: | Mitochondrial distribution and morphology protein 35, Protein UPS1, mitochondrial | | Authors: | Lu, J, Chan, K.C, Zhai, Y, Fan, J, Sun, F. | | Deposit date: | 2016-05-05 | | Release date: | 2017-07-12 | | Last modified: | 2020-09-02 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular mechanism of mitochondrial phosphatidate transfer by Ups1
Commun Biol, 2020
|
|
7KXI
 
 | | Structure of XL5-ligated hRpn13 Pru domain | | Descriptor: | 2-{[(2S)-2-cyano-3-{3-[(4-methylbenzene-1-carbonyl)amino]phenyl}propanoyl]amino}benzoic acid, Proteasomal ubiquitin receptor ADRM1 | | Authors: | Lu, X, Walters, K.J. | | Deposit date: | 2020-12-03 | | Release date: | 2021-12-15 | | Last modified: | 2021-12-29 | | Method: | SOLUTION NMR | | Cite: | Structure-guided bifunctional molecules hit a DEUBAD-lacking hRpn13 species upregulated in multiple myeloma.
Nat Commun, 12, 2021
|
|
7CD6
 
 | | mAPE1-recessed dsDNA product complex | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*AP*C)-3'), DNA-(apurinic or apyrimidinic site) endonuclease | | Authors: | Liu, T.C, Hsiao, Y.Y. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | APE1 distinguishes DNA substrates in exonucleolytic cleavage by induced space-filling.
Nat Commun, 12, 2021
|
|
7CD5
 
 | | mAPE1-blunt-ended dsDNA product complex | | Descriptor: | DNA(5'-D(*CP*GP*TP*AP*AP*TP*AP*CP*G)-3'), DNA-(apurinic or apyrimidinic site) endonuclease | | Authors: | Liu, T.C, Hsiao, Y.Y. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | APE1 distinguishes DNA substrates in exonucleolytic cleavage by induced space-filling.
Nat Commun, 12, 2021
|
|
4HYS
 
 | |
4HYU
 
 | |
