3LFO
 
 | |
3N4Z
 
 | |
3N4Y
 
 | |
3RA6
 
 | |
3RA5
 
 | | Crystal structure of T. celer L30e E6A/R92A variant | | Descriptor: | 50S ribosomal protein L30e, SULFATE ION | | Authors: | Chan, C.H, Wong, K.B. | | Deposit date: | 2011-03-27 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilizing salt-bridge enhances protein thermostability by reducing the heat capacity change of unfolding
Plos One, 6, 2011
|
|
4KQF
 
 | | Crystal structure of CobT E174A complexed with adenine | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase, ... | | Authors: | Chan, C.H, Newmister, S.A, Taylor, K.C, Claas, K.R, Rayment, I, Escalante-Semerena, J.C. | | Deposit date: | 2013-05-15 | | Release date: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dissecting cobamide diversity through structural and functional analyses of the base-activating CobT enzyme of Salmonella enterica.
Biochim.Biophys.Acta, 1840, 2014
|
|
4KQJ
 
 | | Crystal structure of CobT S80Y/Q88M/L175M complexed with p-cresol and NaMN | | Descriptor: | NICOTINATE MONONUCLEOTIDE, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase, P-CRESOL, ... | | Authors: | Chan, C.H, Newmister, S.A, Taylor, K.C, Claas, K.R, Rayment, I, Escalante-Semerena, J.C. | | Deposit date: | 2013-05-15 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dissecting cobamide diversity through structural and functional analyses of the base-activating CobT enzyme of Salmonella enterica.
Biochim.Biophys.Acta, 1840, 2014
|
|
4KQK
 
 | | Crystal structure of CobT S80Y/Q88M/L175M complexed with p-cresol | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase, ... | | Authors: | Chan, C.H, Newmister, S.A, Taylor, K.C, Claas, K.R, Rayment, I, Escalante-Semerena, J.C. | | Deposit date: | 2013-05-15 | | Release date: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Dissecting cobamide diversity through structural and functional analyses of the base-activating CobT enzyme of Salmonella enterica.
Biochim.Biophys.Acta, 1840, 2014
|
|
4KQH
 
 | | Crystal structure of CobT E317A | | Descriptor: | 1,2-ETHANEDIOL, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase, PHOSPHATE ION, ... | | Authors: | Chan, C.H, Newmister, S.A, Taylor, K.C, Claas, K.R, Rayment, I, Escalante-Semerena, J.C. | | Deposit date: | 2013-05-15 | | Release date: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dissecting cobamide diversity through structural and functional analyses of the base-activating CobT enzyme of Salmonella enterica.
Biochim.Biophys.Acta, 1840, 2014
|
|
4KQG
 
 | | Crystal structure of CobT E174A complexed with DMB | | Descriptor: | 1,2-ETHANEDIOL, 5,6-DIMETHYLBENZIMIDAZOLE, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase, ... | | Authors: | Chan, C.H, Newmister, S.A, Taylor, K.C, Claas, K.R, Rayment, I, Escalante-Semerena, J.C. | | Deposit date: | 2013-05-15 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dissecting cobamide diversity through structural and functional analyses of the base-activating CobT enzyme of Salmonella enterica.
Biochim.Biophys.Acta, 1840, 2014
|
|
4KQI
 
 | | Crystal structure of CobT E317A complexed with its reaction products | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-RIBAZOLE-5'-PHOSPHATE, NICOTINIC ACID, ... | | Authors: | Chan, C.H, Newmister, S.A, Taylor, K.C, Claas, K.R, Rayment, I, Escalante-Semerena, J.C. | | Deposit date: | 2013-05-15 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Dissecting cobamide diversity through structural and functional analyses of the base-activating CobT enzyme of Salmonella enterica.
Biochim.Biophys.Acta, 1840, 2014
|
|
8D9M
 
 | | Cryo-EM of the OmcZ nanowires from Geobacter sulfurreducens | | Descriptor: | Cytochrome c, HEME C | | Authors: | Wang, F, Chan, C.H, Mustafa, K, Hochbaum, A.I, Bond, D.R, Egelman, E.H. | | Deposit date: | 2022-06-10 | | Release date: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of Geobacter OmcZ filaments suggests extracellular cytochrome polymers evolved independently multiple times.
Elife, 11, 2022
|
|
7TGG
 
 | | Cryo-EM structure of PilA-N and PilA-C from Geobacter sulfurreducens | | Descriptor: | Geopilin domain 1 protein, Geopilin domain 2 protein | | Authors: | Wang, F, Mustafa, K, Chan, C.H, Joshi, K, Bond, D.R, Hochbaum, A.I, Egelman, E.H. | | Deposit date: | 2022-01-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of an extracellular Geobacter OmcE cytochrome filament reveals tetrahaem packing.
Nat Microbiol, 7, 2022
|
|
7TFS
 
 | | Cryo-EM of the OmcE nanowires from Geobacter sulfurreducens | | Descriptor: | Cytochrome c, HEME C | | Authors: | Wang, F, Mustafa, K, Chan, C.H, Joshi, K, Hochbaum, A.I, Bond, D.R, Egelman, E.H. | | Deposit date: | 2022-01-07 | | Release date: | 2022-05-04 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of an extracellular Geobacter OmcE cytochrome filament reveals tetrahaem packing.
Nat Microbiol, 7, 2022
|
|
4HDM
 
 | | Crystal Structure of ArsAB in Complex with p-cresol | | Descriptor: | 1,2-ETHANEDIOL, ArsA, ArsB, ... | | Authors: | Newmister, S.A, Chan, C.H, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into the Function of the Nicotinate Mononucleotide:phenol/p-cresol Phosphoribosyltransferase (ArsAB) Enzyme from Sporomusa ovata.
Biochemistry, 51, 2012
|
|
4HDN
 
 | | Crystal Structure of ArsAB in the Substrate-Free State. | | Descriptor: | ArsA, ArsB | | Authors: | Newmister, S.A, Chan, C.H, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Insights into the Function of the Nicotinate Mononucleotide:phenol/p-cresol Phosphoribosyltransferase (ArsAB) Enzyme from Sporomusa ovata.
Biochemistry, 51, 2012
|
|
4HDS
 
 | | Crystal Structure of ArsAB in Complex with Phenol. | | Descriptor: | 1,2-ETHANEDIOL, ArsA, ArsB, ... | | Authors: | Newmister, S.A, Chan, C.H, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Function of the Nicotinate Mononucleotide:phenol/p-cresol Phosphoribosyltransferase (ArsAB) Enzyme from Sporomusa ovata.
Biochemistry, 51, 2012
|
|
4HDK
 
 | | Crystal Structure of ArsAB in Complex with Phloroglucinol | | Descriptor: | 1,2-ETHANEDIOL, ArsA, ArsB, ... | | Authors: | Newmister, S.A, Chan, C.H, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Function of the Nicotinate Mononucleotide:phenol/p-cresol Phosphoribosyltransferase (ArsAB) Enzyme from Sporomusa ovata.
Biochemistry, 51, 2012
|
|
4HDR
 
 | | Crystal Structure of ArsAB in Complex with 5,6-dimethylbenzimidazole | | Descriptor: | 1,2-ETHANEDIOL, 5,6-DIMETHYLBENZIMIDAZOLE, ArsA, ... | | Authors: | Newmister, S.A, Chan, C.H, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Insights into the Function of the Nicotinate Mononucleotide:phenol/p-cresol Phosphoribosyltransferase (ArsAB) Enzyme from Sporomusa ovata.
Biochemistry, 51, 2012
|
|
7ASW
 
 | | Crystal structure of chloroplastic thioredoxin z defines a novel type-specific target recognition | | Descriptor: | Thioredoxin-related protein CITRX | | Authors: | Le Moigne, T, Gurrieri, L, Crozet, P, Marchand, C.H, Zaffagnini, M, Sparla, F, Lemaire, S.D, Henri, J. | | Deposit date: | 2020-10-28 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Crystal structure of chloroplastic thioredoxin z defines a type-specific target recognition.
Plant J., 107, 2021
|
|
1BWB
 
 | | HIV-1 PROTEASE (V82F/I84V) DOUBLE MUTANT COMPLEXED WITH SD146 OF DUPONT PHARMACEUTICALS | | Descriptor: | PROTEIN (HIV-1 PROTEASE), [4R-(4ALPHA,5ALPHA,6ALPHA,7ALPHA)]-3,3'-{{TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-DIYL]BIS(METHYLENE)]BIS[N-1H-BENZIMIDAZOL-2-YLBENZAMIDE] | | Authors: | Ala, P, Chang, C.H. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities.
Biochemistry, 37, 1998
|
|
1BV7
 
 | | COUNTERACTING HIV-1 PROTEASE DRUG RESISTANCE: STRUCTURAL ANALYSIS OF MUTANT PROTEASES COMPLEXED WITH XV638 AND SD146, CYCLIC UREA AMIDES WITH BROAD SPECIFICITIES | | Descriptor: | PROTEIN (HIV-1 PROTEASE), [4R-(4ALPHA,5ALPHA,6BETA,7BETA)]-3,3'-[[TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-D IYL] BIS(METHYLENE)]BIS[N-2-THIAZOLYLBENZAMIDE] | | Authors: | Ala, P, Chang, C.H. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities.
Biochemistry, 37, 1998
|
|
1BWA
 
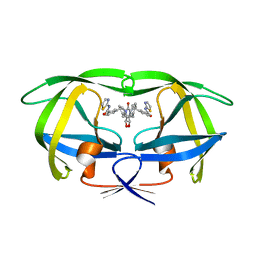 | | HIV-1 PROTEASE (V82F/I84V) DOUBLE MUTANT COMPLEXED WITH XV638 OF DUPONT PHARMACEUTICALS | | Descriptor: | PROTEIN (HIV-1 PROTEASE), [4R-(4ALPHA,5ALPHA,6BETA,7BETA)]-3,3'-[[TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-D IYL] BIS(METHYLENE)]BIS[N-2-THIAZOLYLBENZAMIDE] | | Authors: | Ala, P, Chang, C.H. | | Deposit date: | 1998-09-22 | | Release date: | 1998-10-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities.
Biochemistry, 37, 1998
|
|
1BV9
 
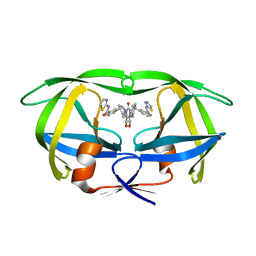 | | HIV-1 PROTEASE (I84V) COMPLEXED WITH XV638 OF DUPONT PHARMACEUTICALS | | Descriptor: | PROTEIN (HIV-1 PROTEASE), [4R-(4ALPHA,5ALPHA,6BETA,7BETA)]-3,3'-[[TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-D IYL] BIS(METHYLENE)]BIS[N-2-THIAZOLYLBENZAMIDE] | | Authors: | Ala, P, Chang, C.H. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities.
Biochemistry, 37, 1998
|
|
1WTL
 
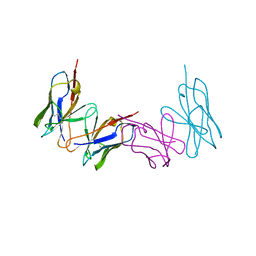 | |
