5HL1
 
 | | Crystal structure of glutaminase C in complex with inhibitor CB-839 | | Descriptor: | 2-(pyridin-2-yl)-N-(5-{4-[6-({[3-(trifluoromethoxy)phenyl]acetyl}amino)pyridazin-3-yl]butyl}-1,3,4-thiadiazol-2-yl)acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2016-01-14 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the clinically relevant glutaminase inhibitot CB-839 in complex with glutaminase C
To Be Published
|
|
5WJ6
 
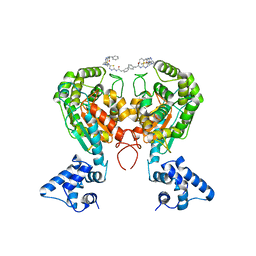 | | Crystal structure of glutaminase C in complex with inhibitor 2-phenyl-N-{5-[4-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}amino)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide (UPGL-00004) | | Descriptor: | 2-phenyl-N-{5-[4-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}amino)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Characterization of the interactions of potent allosteric inhibitors with glutaminase C, a key enzyme in cancer cell glutamine metabolism.
J. Biol. Chem., 293, 2018
|
|
3FEY
 
 | |
3FEX
 
 | |
1GRN
 
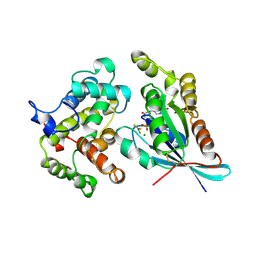 | | CRYSTAL STRUCTURE OF THE CDC42/CDC42GAP/ALF3 COMPLEX. | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nassar, N, Hoffman, G.R, Clardy, J.C, Cerione, R.A. | | Deposit date: | 1998-07-30 | | Release date: | 1999-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP.
Nat.Struct.Biol., 5, 1998
|
|
1KV3
 
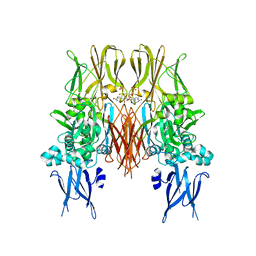 | | HUMAN TISSUE TRANSGLUTAMINASE IN GDP BOUND FORM | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Protein-glutamine gamma-glutamyltransferase | | Authors: | Liu, S, Cerione, R.A, Clardy, J. | | Deposit date: | 2002-01-24 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the guanine nucleotide-binding activity of tissue transglutaminase and its regulation of transamidation activity.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2GCN
 
 | |
7SBN
 
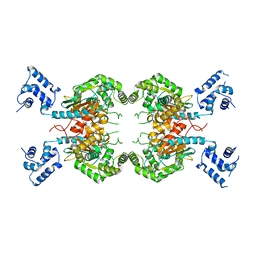 | |
7SBM
 
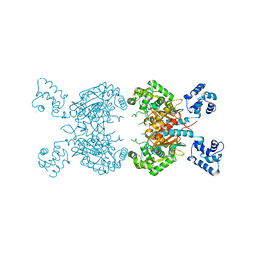 | | Human glutaminase C (Y466W) with L-Gln, open conformation | | Descriptor: | GLUTAMINE, Isoform 3 of Glutaminase kidney isoform, mitochondrial | | Authors: | Nguyen, T.-T.T, Cerione, R.A. | | Deposit date: | 2021-09-25 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High-resolution structures of mitochondrial glutaminase C tetramers indicate conformational changes upon phosphate binding.
J.Biol.Chem., 298, 2022
|
|
2GCO
 
 | | Crystal structure of the human RhoC-GppNHp complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Rho-related GTP-binding protein RhoC | | Authors: | Dias, S.M.G, Cerione, R.A. | | Deposit date: | 2006-03-14 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray Crystal Structures Reveal Two Activated States for RhoC.
Biochemistry, 46, 2007
|
|
2GCP
 
 | | Crystal structure of the human RhoC-GSP complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Dias, S.M.G, Cerione, R.A. | | Deposit date: | 2006-03-14 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray Crystal Structures Reveal Two Activated States for RhoC.
Biochemistry, 46, 2007
|
|
1NI2
 
 | | Structure of the active FERM domain of Ezrin | | Descriptor: | Ezrin | | Authors: | Smith, W.J, Nassar, N, Bretscher, A.P, Cerione, R.A, Karplus, P.A. | | Deposit date: | 2002-12-20 | | Release date: | 2003-02-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Active N-terminal Domain of Ezrin. CONFORMATIONAL AND MOBILITY CHANGES IDENTIFY KEYSTONE INTERACTIONS.
J.Biol.Chem., 278, 2003
|
|
1PZD
 
 | | Structural Identification of a conserved appendage domain in the carboxyl-terminus of the COPI gamma-subunit. | | Descriptor: | Coatomer gamma subunit, SULFATE ION | | Authors: | Hoffman, G.R, Rahl, P.B, Collins, R.N, Cerione, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Conserved Structural Motifs in Intracellular Trafficking Pathways. Structure of the gammaCOP Appendage Domain.
Mol.Cell, 12, 2003
|
|
7RGG
 
 | | Room temperature serial crystal structure of Glutaminase C in complex with inhibitor BPTES | | Descriptor: | Glutaminase kidney isoform, mitochondrial 68 kDa chain, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide) | | Authors: | Milano, S.K, Finke, A, Cerione, R.A. | | Deposit date: | 2021-07-15 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New insights into the molecular mechanisms of glutaminase C inhibitors in cancer cells using serial room temperature crystallography.
J.Biol.Chem., 298, 2022
|
|
7REN
 
 | | Room temperature serial crystal structure of Glutaminase C in complex with inhibitor UPGL-00004 | | Descriptor: | 2-phenyl-N-{5-[4-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}amino)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Milano, S.K, Finke, A, Cerione, R.A. | | Deposit date: | 2021-07-13 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | New insights into the molecular mechanisms of glutaminase C inhibitors in cancer cells using serial room temperature crystallography.
J.Biol.Chem., 298, 2022
|
|
1AN0
 
 | | CDC42HS-GDP COMPLEX | | Descriptor: | CDC42HS-GDP, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Kongsaeree, P, Cerione, R, Clardy, J. | | Deposit date: | 1997-06-26 | | Release date: | 1999-01-13 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure Determination of Cdc42Hs and Gdp Complex
To be Published
|
|
2NGR
 
 | | TRANSITION STATE COMPLEX FOR GTP HYDROLYSIS BY CDC42: COMPARISONS OF THE HIGH RESOLUTION STRUCTURES FOR CDC42 BOUND TO THE ACTIVE AND CATALYTICALLY COMPROMISED FORMS OF THE CDC42-GAP. | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nassar, N, Hoffman, G, Clardy, J, Cerione, R. | | Deposit date: | 1998-07-31 | | Release date: | 1999-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP.
Nat.Struct.Biol., 5, 1998
|
|
4F4U
 
 | |
4F56
 
 | | The bicyclic intermediate structure provides insights into the desuccinylation mechanism of SIRT5 | | Descriptor: | 3-[(2R,3aR,5R,6R,6aR)-5-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-2,6-dihydroxytetrahydrofuro[2,3-d][1,3]oxathiol-2-yl]propanoic acid, NAD-dependent lysine demalonylase and desuccinylase sirtuin-5, mitochondrial, ... | | Authors: | Zhou, Y, Hao, Q. | | Deposit date: | 2012-05-11 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Bicyclic Intermediate Structure Provides Insights into the Desuccinylation Mechanism of Human Sirtuin 5 (SIRT5)
J.Biol.Chem., 287, 2012
|
|
1AJW
 
 | |
3SS4
 
 | |
3SS3
 
 | |
1DOA
 
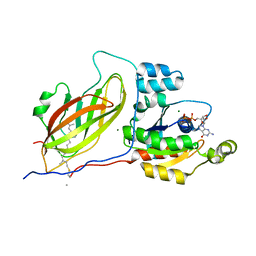 | | Structure of the rho family gtp-binding protein cdc42 in complex with the multifunctional regulator rhogdi | | Descriptor: | GERAN-8-YL GERAN, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hoffman, G.R, Nassar, N, Cerione, R.C. | | Deposit date: | 1999-12-20 | | Release date: | 2000-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI.
Cell(Cambridge,Mass.), 100, 2000
|
|
3RIG
 
 | |
3RIY
 
 | |
