6ZXA
 
 | | LH2 complex from Marichromatium purpuratum | | Descriptor: | 9-cis-okenone, BACTERIOCHLOROPHYLL A, LHC domain-containing protein, ... | | Authors: | Gardiner, A.T, Naydenova, K, Castro-Hartmann, P, Nguyen-Phan, T.C, Russo, C.J, Sader, K, Hunter, C.N, Cogdell, R.J, Qian, P. | | Deposit date: | 2020-07-29 | | Release date: | 2021-02-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | The 2.4 angstrom cryo-EM structure of a heptameric light-harvesting 2 complex reveals two carotenoid energy transfer pathways.
Sci Adv, 7, 2021
|
|
7WLM
 
 | | The Cryo-EM structure of siphonaxanthin chlorophyll a/b type light-harvesting complex II | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, CHLOROPHYLL A, ... | | Authors: | Seki, S, Nakaniwa, T, Castro-Hartmann, P, Sader, K, Kawamoto, A, Tanaka, H, Qian, P, Kurisu, G, Fujii, R. | | Deposit date: | 2022-01-13 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into blue-green light utilization by marine green algal light harvesting complex II at 2.78 angstrom.
Bba Adv, 2, 2022
|
|
7PBW
 
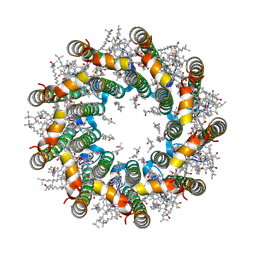 | | Cryo-EM structure of light harvesting complex 2 from Rba. sphaeroides. | | Descriptor: | BACTERIOCHLOROPHYLL A, CALCIUM ION, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Qian, P, Swainsbury, D.J.K, Croll, T.I, Castro-Hartmann, P, Sader, K, Divitini, G, Hunter, C.N. | | Deposit date: | 2021-08-02 | | Release date: | 2021-11-24 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Cryo-EM Structure of the Rhodobacter sphaeroides Light-Harvesting 2 Complex at 2.1 angstrom.
Biochemistry, 60, 2021
|
|
7PHL
 
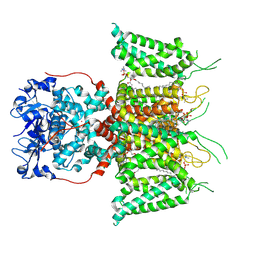 | | Human voltage-gated potassium channel Kv3.1 (with EDTA) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, POTASSIUM ION, Potassium voltage-gated channel, ... | | Authors: | Chi, G, Qian, P, Castro-Hartmann, P, Venkaya, S, Singh, N.K, McKinley, G, Mukhopadhyay, S.M.M, Fernandez-Cid, A, Pike, A.C.W, Marsden, B, MacLean, E.M, Sader, K, Burgess-Brown, N.A, Duerr, K.L. | | Deposit date: | 2021-08-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the human Kv3.1 channel reveals gating control by the cytoplasmic T1 domain.
Nat Commun, 13, 2022
|
|
7PHI
 
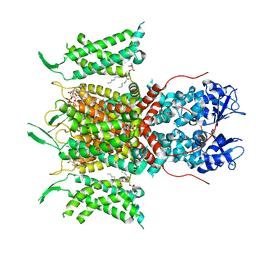 | | Human voltage-gated potassium channel Kv3.1 (with Zn) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, POTASSIUM ION, Potassium voltage-gated channel, ... | | Authors: | Chi, G, Qian, P, Castro-Hartmann, P, Venkaya, S, Singh, N.K, McKinley, G, Mukhopadhyay, S.M.M, Fernandez-Cid, A, Marsden, B, MacLean, E.M, Pike, A.C.W, Sader, K, Burgess-Brown, N.A, Duerr, K.L. | | Deposit date: | 2021-08-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the human Kv3.1 channel reveals gating control by the cytoplasmic T1 domain.
Nat Commun, 13, 2022
|
|
7PHK
 
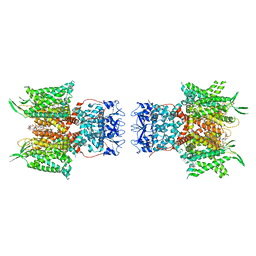 | | Human voltage-gated potassium channel Kv3.1 in dimeric state (with Zn) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, POTASSIUM ION, Potassium voltage-gated channel, ... | | Authors: | Chi, G, Qian, P, Castro-Hartmann, P, Venkaya, S, Singh, N.K, McKinley, G, Mukhopadhyay, S.M.M, Marsden, B, MacLean, E.M, Fernandez-Cid, A, Pike, A.C.W, Sader, K, Burgess-Brown, N.A, Duerr, K.L. | | Deposit date: | 2021-08-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the human Kv3.1 channel reveals gating control by the cytoplasmic T1 domain.
Nat Commun, 13, 2022
|
|
7AAP
 
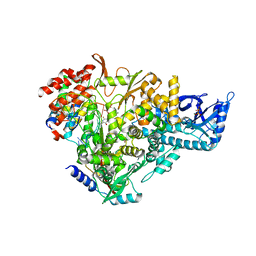 | | Nsp7-Nsp8-Nsp12 SARS-CoV2 RNA-dependent RNA polymerase in complex with template:primer dsRNA and favipiravir-RTP | | Descriptor: | MAGNESIUM ION, Non-structural protein 12, Non-structural protein 7, ... | | Authors: | Naydenova, K, Muir, K.W, Wu, L.F, Zhang, Z, Coscia, F, Peet, M, Castro-Hartman, P, Qian, P, Sader, K, Dent, K, Kimanius, D, Sutherland, J.D, Lowe, J, Barford, D, Russo, C.J. | | Deposit date: | 2020-09-04 | | Release date: | 2020-09-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OY8
 
 | | Cryo-EM structure of the Rhodospirillum rubrum RC-LH1 complex | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, 2-azanyl-5-[(2~{E},6~{E},8~{E},10~{E},12~{E},14~{E},18~{E},22~{E},26~{E},30~{E},34~{E})-3,7,11,15,19,23,27,31,35,39-decamethyltetraconta-2,6,8,10,12,14,18,22,26,30,34,38-dodecaenyl]-3-methoxy-6-methyl-cyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Qian, P, Croll, T.I, Castro, H.P, Moriarty, N.W, sader, K, Hunter, C.N. | | Deposit date: | 2021-06-23 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of the Rhodospirillum rubrum RC-LH1 complex at 2.5 angstrom.
Biochem.J., 478, 2021
|
|
7K0S
 
 | |
7UMZ
 
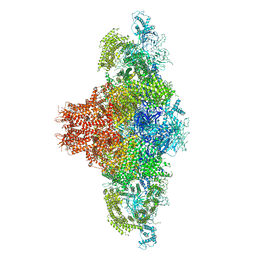 | |
7O0W
 
 | | Cryo-EM structure of the RC-dLH complex (model_1b) from Gemmatimonas phototrophica at 2.47 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0U
 
 | | Cryo-EM structure (model_1a) of the RC-dLH complex from Gemmatimonas phototrophica at 2.4 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0V
 
 | | Cryo-EM structure (model_2a) of the RC-dLH complex from Gemmatimonas phototrophica at 2.5 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0X
 
 | | Cryo-EM structure (model_2b) of the RC-dLH complex from Gemmatimonas phototrophica at 2.44 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-28 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7ZDI
 
 | | PucB-LH2 complex from Rps. palustris | | Descriptor: | 1,2-Dihydro-psi,psi-caroten-1-ol, BACTERIOCHLOROPHYLL A, Light-harvesting protein, ... | | Authors: | Qian, P, Cogdell, R.J, Nguyen-Phan, T.C. | | Deposit date: | 2022-03-29 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of light-harvesting 2 complexes from Rhodopseudomonas palustris reveal the molecular origin of absorption tuning.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZE3
 
 | | PucD-LH2 complex from Rps. palustris | | Descriptor: | (3'E)-3',4'-didehydro-1,2-dihydro-psi,psi-caroten-1-ol, BACTERIOCHLOROPHYLL A, Light-harvesting protein B-800-850 alpha chain, ... | | Authors: | Qian, P, Cogdell, R.J, Nguyen-Phan, T.C. | | Deposit date: | 2022-03-30 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of light-harvesting 2 complexes from Rhodopseudomonas palustris reveal the molecular origin of absorption tuning.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZE8
 
 | | PucE-LH2 complex from Rps. palustris | | Descriptor: | 1,2-Dihydro-psi,psi-caroten-1-ol, BACTERIOCHLOROPHYLL A, Light-harvesting protein, ... | | Authors: | Qian, P, Cogdell, R.J, Nguyen-Phan, T.C. | | Deposit date: | 2022-03-30 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of light-harvesting 2 complexes from Rhodopseudomonas palustris reveal the molecular origin of absorption tuning.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZCU
 
 | | PucA-LH2 complex from Rps. palustris | | Descriptor: | 1,2-Dihydro-psi,psi-caroten-1-ol, BACTERIOCHLOROPHYLL A, Light-harvesting protein B-800-850 alpha chain, ... | | Authors: | Qian, P, Cogdell, R.J. | | Deposit date: | 2022-03-28 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of light-harvesting 2 complexes from Rhodopseudomonas palustris reveal the molecular origin of absorption tuning.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6QIK
 
 | |
6QT0
 
 | |
6QTZ
 
 | |
7PIL
 
 | | Cryo-EM structure of the Rhodobacter sphaeroides RC-LH1-PufXY monomer complex at 2.5 A | | Descriptor: | (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, BACTERIOCHLOROPHYLL A, ... | | Authors: | Qian, P, Hunter, C.N. | | Deposit date: | 2021-08-20 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of the monomeric Rhodobacter sphaeroides RC-LH1 core complex at 2.5 angstrom.
Biochem.J., 478, 2021
|
|
7PQD
 
 | | Cryo-EM structure of the dimeric Rhodobacter sphaeroides RC-LH1 core complex at 2.9 A: the structural basis for dimerisation | | Descriptor: | (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Qian, P, Hunter, C.N. | | Deposit date: | 2021-09-17 | | Release date: | 2021-11-24 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of the dimeric Rhodobacter sphaeroides RC-LH1 core complex at 2.9 angstrom : the structural basis for dimerisation.
Biochem.J., 478, 2021
|
|
7PHH
 
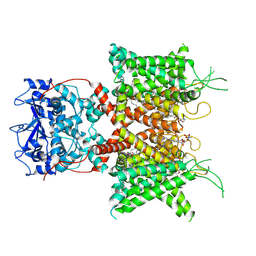 | | Human voltage-gated potassium channel Kv3.1 (apo condition) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, POTASSIUM ION, Potassium voltage-gated channel, ... | | Authors: | Chi, G, Venkaya, S, Singh, N.K, McKinley, G, Mukhopadhyay, S.M.M, Marsden, B, MacLean, E.M, Fernandez-Cid, A, Pike, A.C.W, Savva, C, Ragan, T.J, Burgess-Brown, N.A, Duerr, K.L. | | Deposit date: | 2021-08-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the human Kv3.1 channel reveals gating control by the cytoplasmic T1 domain.
Nat Commun, 13, 2022
|
|
6RI5
 
 | |
