8I3U
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 14B1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 14B1, Light chain of Fab 14B1, ... | | Authors: | Li, Z, Yu, F, Cao, S. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
8I3S
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 7B3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain od Fab 7B3, Light chain of Fab 7B3, ... | | Authors: | Li, Z, Yu, F, Cao, S, ZHao, H. | | Deposit date: | 2023-01-17 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
2MAI
 
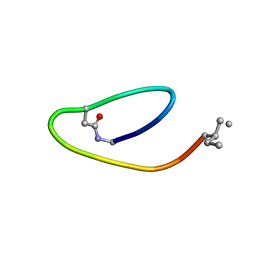 | | NMR structure of lassomycin | | Descriptor: | Lassomycin | | Authors: | Gavrish, E, Sit, C.S, Kandror, O, Spoering, A, Peoples, A, Ling, L, Fetterman, A, Hughes, D, Cao, S, Bissell, A, Torrey, H, Akopian, T, Mueller, A, Epstein, S, Goldberg, A, Clardy, J, Lewis, K. | | Deposit date: | 2013-07-09 | | Release date: | 2014-05-07 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Lassomycin, a Ribosomally Synthesized Cyclic Peptide, Kills Mycobacterium tuberculosis by Targeting the ATP-Dependent Protease ClpC1P1P2.
Chem.Biol., 21, 2014
|
|
7FGF
 
 | |
7YNY
 
 | |
7EJ8
 
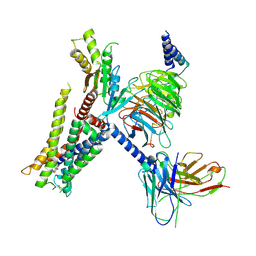 | | Structure of the alpha2A-adrenergic receptor GoA signaling complex bound to brimonidine | | Descriptor: | Alpha-2A adrenergic receptor, Brimonidine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Cao, S, Liu, Z, Du, Y. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into ligand recognition, activation, and signaling of the alpha 2A adrenergic receptor.
Sci Adv, 8, 2022
|
|
7EJ0
 
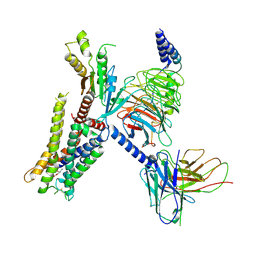 | | Structure of the alpha2A-adrenergic receptor GoA signaling complex | | Descriptor: | Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, J, Cao, S, Liu, Z, Du, Y. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into ligand recognition, activation, and signaling of the alpha 2A adrenergic receptor.
Sci Adv, 8, 2022
|
|
7EJA
 
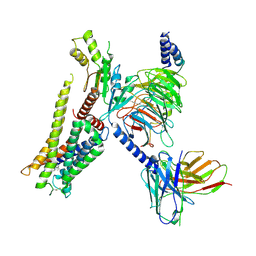 | | Structure of the alpha2A-adrenergic receptor GoA signaling complex bound to dexmedetomidine | | Descriptor: | 4-[(1~{S})-1-(2,3-dimethylphenyl)ethyl]-1~{H}-imidazole, Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Cao, S, Liu, Z, Du, Y. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into ligand recognition, activation, and signaling of the alpha 2A adrenergic receptor.
Sci Adv, 8, 2022
|
|
7EJK
 
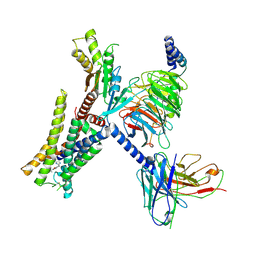 | | Structure of the alpha2A-adrenergic receptor GoA signaling complex bound to oxymetazoline | | Descriptor: | Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, J, Cao, S, Liu, Z, Du, Y. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into ligand recognition, activation, and signaling of the alpha 2A adrenergic receptor.
Sci Adv, 8, 2022
|
|
8GYN
 
 | | zebrafish TIPE1 strucutre in complex with PE | | Descriptor: | Tumor necrosis factor alpha-induced protein 8-like protein 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Wang, W, Cao, S.J. | | Deposit date: | 2022-09-23 | | Release date: | 2023-04-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural insight into TIPE1 functioning as a lipid transfer protein.
J.Biomol.Struct.Dyn., 41, 2023
|
|
7P2M
 
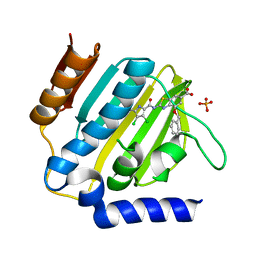 | | E.coli GyrB24 with inhibitor LMD43 (EBL2560) | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-phenylmethoxy-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7P2W
 
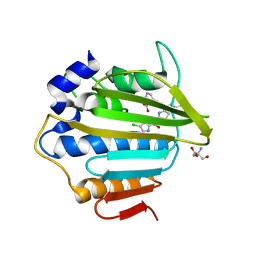 | | E.coli GyrB24 with inhibitor LMD92 (EBL2682) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(3-carboxyphenyl)methoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, ... | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
6Q0C
 
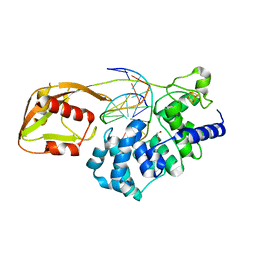 | | MutY adenine glycosylase bound to DNA containing a transition state analog (1N) paired with undamaged dG | | Descriptor: | 1,2-ETHANEDIOL, A/G-specific adenine glycosylase, CALCIUM ION, ... | | Authors: | O'Shea Murray, V.L, Russelburg, L.P, Horvath, M.P, David, S.S. | | Deposit date: | 2019-08-01 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Finding OG Lesions and Avoiding Undamaged G by the DNA Glycosylase MutY.
Acs Chem.Biol., 15, 2020
|
|
5KU9
 
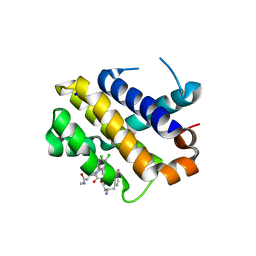 | | Crystal structure of MCL1 with compound 1 | | Descriptor: | (3~{S})-3-azanyl-4-(4-bromophenyl)-~{N}-[(3~{S})-1-[2-[[(2~{R})-1-(3,4-dichlorophenyl)-4-(methylamino)-4-oxidanylidene-butan-2-yl]amino]-2-oxidanylidene-ethyl]-2-oxidanylidene-4,5-dihydro-3~{H}-1-benzazepin-3-yl]butanamide, Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-07-13 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5MES
 
 | | MCL1 FAB COMPLEX IN COMPLEX WITH COMPOUND 29 | | Descriptor: | (5~{R},13~{S},17~{S})-5-[[4-chloranyl-3-(2-phenylethyl)phenyl]methyl]-13-[(4-chlorophenyl)methyl]-8-methyl-1,4,8,12,16-pentazatricyclo[15.8.1.0^{20,25}]hexacosa-20(25),21,23-triene-3,7,15,26-tetrone, Heavy Chain, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog,Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Hargreaves, D. | | Deposit date: | 2016-11-16 | | Release date: | 2017-01-18 | | Last modified: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5MEV
 
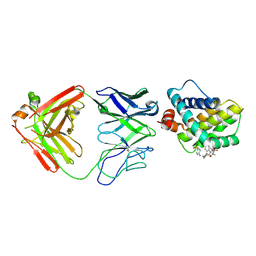 | | MCL1 FAB COMPLEX IN COMPLEX WITH COMPOUND 21 | | Descriptor: | (5~{R},13~{S},17~{S})-5-[(3,4-dichlorophenyl)methyl]-8-methyl-13-[(4-methylsulfonylphenyl)methyl]-1,4,8,12,16-pentazatricyclo[15.8.1.0^{20,25}]hexacosa-20,22,24-triene-3,7,15,26-tetrone, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Hargreaves, D. | | Deposit date: | 2016-11-16 | | Release date: | 2017-01-18 | | Last modified: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors
Acs Med.Chem.Lett., 8, 2017
|
|
8QK5
 
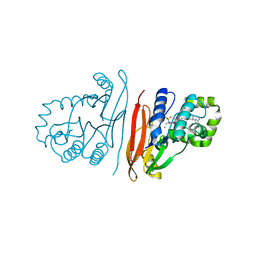 | | Structure of K. pneumoniae LpxH in complex with EBL-3647 | | Descriptor: | 5-[[3-(aminomethyl)azetidin-1-yl]methyl]-N-[4-[4-(4-cyano-6-methyl-pyrimidin-2-yl)piperazin-1-yl]sulfonylphenyl]-2-[methyl(methylsulfonyl)amino]benzamide, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QJZ
 
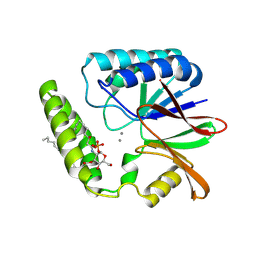 | | Crystal structure of E. coli LpxH in complex with lipid X | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QK2
 
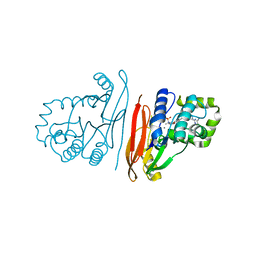 | | Structure of K.pneumoniae LpxH in complex with EBL-3339 | | Descriptor: | MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase, ~{N}-[4-[4-(4-cyano-6-methyl-pyrimidin-2-yl)piperazin-1-yl]sulfonylphenyl]-2-[methyl(methylsulfonyl)amino]benzamide | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QK9
 
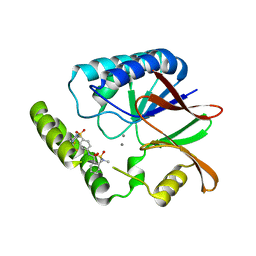 | | Structure of E. coli LpxH in complex with JEDI-1444 | | Descriptor: | 2-[methyl(methylsulfonyl)amino]-~{N}-[4-[4-[3-(trifluoromethyl)phenyl]piperazin-1-yl]sulfonylphenyl]benzamide, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QKA
 
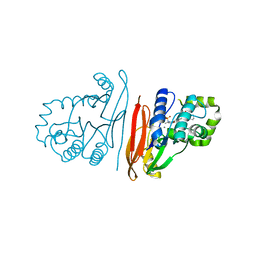 | | Structure of K. pneumoniae LpxH in complex with JEDI-852 | | Descriptor: | 2-[methyl(methylsulfonyl)amino]-~{N}-(4-piperidin-1-ylsulfonylphenyl)benzamide, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7KH0
 
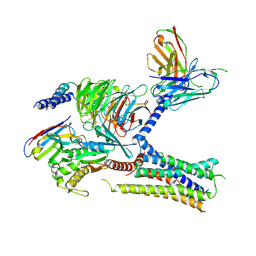 | | Cryo-EM structure of the human arginine vasopressin AVP-vasopressin receptor V2R-Gs signaling complex | | Descriptor: | Arg-vasopressin, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, L, Xu, J, Gao, S, Sun, D, Liu, H, Liu, Z, Du, Y, Zhang, C. | | Deposit date: | 2020-10-19 | | Release date: | 2021-05-26 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of the AVP-vasopressin receptor 2-G s signaling complex.
Cell Res., 31, 2021
|
|
8BN6
 
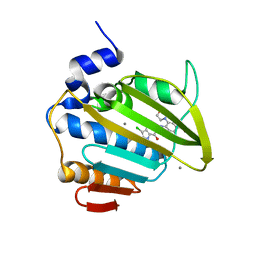 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with EBL3021 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-morpholin-4-yl-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, DNA gyrase subunit B | | Authors: | Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2022-11-12 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Dual Inhibitors of Bacterial Topoisomerases with Broad-Spectrum Antibacterial Activity and In Vivo Efficacy against Vancomycin-Intermediate Staphylococcus aureus .
J.Med.Chem., 66, 2023
|
|
7PTF
 
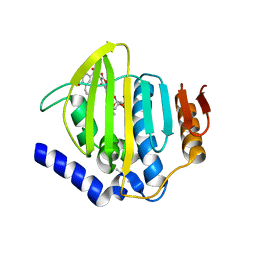 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with novobiocin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA gyrase subunit B, ... | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PQL
 
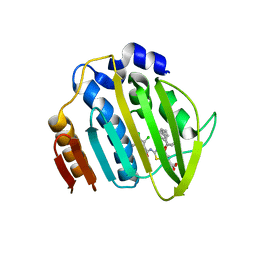 | | Acinetobacter baumannii DNA gyrase B 23kDa ATPase subdomain complexed with EBL2704 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1R)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1S)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
