8QKA
 
 | | Structure of K. pneumoniae LpxH in complex with JEDI-852 | | Descriptor: | 2-[methyl(methylsulfonyl)amino]-~{N}-(4-piperidin-1-ylsulfonylphenyl)benzamide, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QK9
 
 | | Structure of E. coli LpxH in complex with JEDI-1444 | | Descriptor: | 2-[methyl(methylsulfonyl)amino]-~{N}-[4-[4-[3-(trifluoromethyl)phenyl]piperazin-1-yl]sulfonylphenyl]benzamide, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5KU9
 
 | | Crystal structure of MCL1 with compound 1 | | Descriptor: | (3~{S})-3-azanyl-4-(4-bromophenyl)-~{N}-[(3~{S})-1-[2-[[(2~{R})-1-(3,4-dichlorophenyl)-4-(methylamino)-4-oxidanylidene-butan-2-yl]amino]-2-oxidanylidene-ethyl]-2-oxidanylidene-4,5-dihydro-3~{H}-1-benzazepin-3-yl]butanamide, Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-07-13 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5MES
 
 | | MCL1 FAB COMPLEX IN COMPLEX WITH COMPOUND 29 | | Descriptor: | (5~{R},13~{S},17~{S})-5-[[4-chloranyl-3-(2-phenylethyl)phenyl]methyl]-13-[(4-chlorophenyl)methyl]-8-methyl-1,4,8,12,16-pentazatricyclo[15.8.1.0^{20,25}]hexacosa-20(25),21,23-triene-3,7,15,26-tetrone, Heavy Chain, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog,Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Hargreaves, D. | | Deposit date: | 2016-11-16 | | Release date: | 2017-01-18 | | Last modified: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5MEV
 
 | | MCL1 FAB COMPLEX IN COMPLEX WITH COMPOUND 21 | | Descriptor: | (5~{R},13~{S},17~{S})-5-[(3,4-dichlorophenyl)methyl]-8-methyl-13-[(4-methylsulfonylphenyl)methyl]-1,4,8,12,16-pentazatricyclo[15.8.1.0^{20,25}]hexacosa-20,22,24-triene-3,7,15,26-tetrone, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Hargreaves, D. | | Deposit date: | 2016-11-16 | | Release date: | 2017-01-18 | | Last modified: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors
Acs Med.Chem.Lett., 8, 2017
|
|
7KH0
 
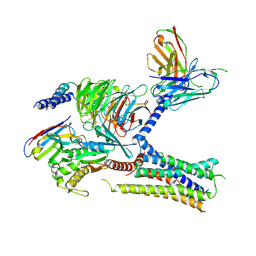 | | Cryo-EM structure of the human arginine vasopressin AVP-vasopressin receptor V2R-Gs signaling complex | | Descriptor: | Arg-vasopressin, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, L, Xu, J, Gao, S, Sun, D, Liu, H, Liu, Z, Du, Y, Zhang, C. | | Deposit date: | 2020-10-19 | | Release date: | 2021-05-26 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of the AVP-vasopressin receptor 2-G s signaling complex.
Cell Res., 31, 2021
|
|
7BQK
 
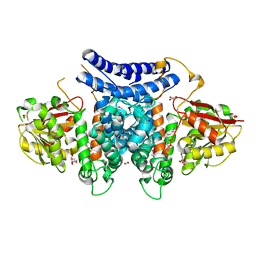 | | The structure of PdxI in complex with its substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, 3-[(E,2S,4S)-2,4-dimethyloct-6-enoyl]-4-oxidanyl-1H-pyridin-2-one, GLYCEROL, ... | | Authors: | Cai, Y.J, Ohashi, M, Zhou, J.H, Tang, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | An enzymatic Alder-ene reaction.
Nature, 586, 2020
|
|
7BQL
 
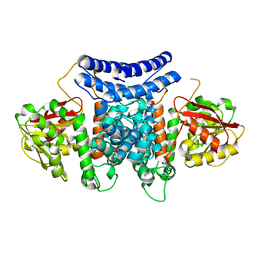 | | The crystal structure of PdxI complex with the Alder-ene adduct | | Descriptor: | 3-[(1R,2S,4R,6S)-2-ethenyl-4,6-dimethyl-cyclohexyl]-4-oxidanyl-1H-pyridin-2-one, DI(HYDROXYETHYL)ETHER, Methyltransf_2 domain-containing protein | | Authors: | Cai, Y.J, Ohashi, M, Zhou, J.H, Tang, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | An enzymatic Alder-ene reaction.
Nature, 586, 2020
|
|
3CTM
 
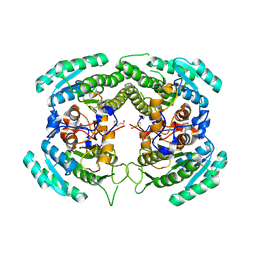 | | Crystal Structure of a Carbonyl Reductase from Candida Parapsilosis with anti-Prelog Stereo-specificity | | Descriptor: | Carbonyl Reductase | | Authors: | Zhang, R, Zhu, G, Li, X, Xu, Y, Zhang, X.C, Rao, Z. | | Deposit date: | 2008-04-14 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of a carbonyl reductase from Candida parapsilosis with anti-Prelog stereospecificity.
Protein Sci., 17, 2008
|
|
1KU5
 
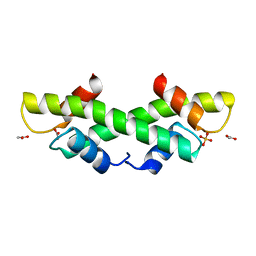 | | Crystal Structure of recombinant histone HPhA from hyperthermophilic archaeon Pyrococcus horikoshii OT3 | | Descriptor: | ACETATE ION, HPhA, SULFATE ION | | Authors: | Li, T, Sun, F, Ji, X, Feng, Y, Rao, Z. | | Deposit date: | 2002-01-21 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure based hyperthermostability of archaeal histone HPhA from Pyrococcus horikoshii
J.MOL.BIOL., 325, 2003
|
|
7T2Z
 
 | |
7T33
 
 | |
4RNA
 
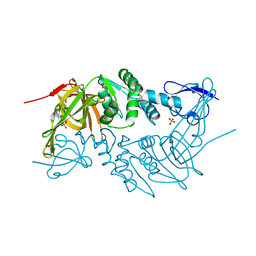 | | Crystal structure of PLpro from Middle East Respiratory Syndrome (MERS) coronavirus | | Descriptor: | PHOSPHATE ION, ZINC ION, papain-like protease | | Authors: | Lei, H, Santarsiero, B.D, Lee, H, Johnson, M.E. | | Deposit date: | 2014-10-23 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Inhibitor Recognition Specificity of MERS-CoV Papain-like Protease May Differ from That of SARS-CoV.
Acs Chem.Biol., 10, 2015
|
|
8GDA
 
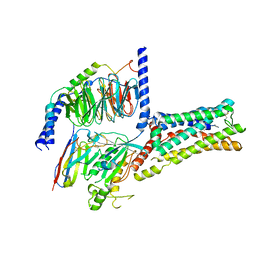 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein | | Descriptor: | (2S,5R)-5-[(1E,3S)-4,4-difluoro-3-hydroxy-4-phenylbut-1-en-1-yl]-1-[6-(1H-tetrazol-5-yl)hexyl]pyrrolidin-2-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Shenming, H, Mengyao, X, Lei, L, Jiangqian, M, Sheng, C, Jinpeng, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GDB
 
 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Shenming, H, Mengyao, X, Lei, L, Jianqiang, M, Jinpeng, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GDC
 
 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 3 Coupled to G Protein | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Shenming, H, Mengyao, X, Lei, L, Yang, D, Shiyi, G, Jinpeng, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GD9
 
 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein | | Descriptor: | 4-({2-[(1R,2R,3R,5S)-3,5-dihydroxy-2-{(3S)-3-hydroxy-4-[3-(methoxymethyl)phenyl]butyl}cyclopentyl]ethyl}sulfanyl)butanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Shenming, H, Mengyao, X, Lei, L, Yang, D, Sheng, C, Jinpeng, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GCM
 
 | | Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein | | Descriptor: | (5S)-5-[(3R)-4,4-difluoro-3-hydroxy-4-phenylbutyl]-1-[6-(1H-tetrazol-5-yl)hexyl]pyrrolidin-2-one, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Huang, S.M, Xiong, M.Y, Liu, L, Mu, J, Sheng, C, Sun, J. | | Deposit date: | 2023-03-02 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GCP
 
 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Huang, S.M, Xiong, M.Y, Liu, L, Mu, J, Sheng, C, Sun, J. | | Deposit date: | 2023-03-02 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8H97
 
 | | GH86 agarase Aga86A_Wa | | Descriptor: | Beta-agarase, CALCIUM ION, HEXAETHYLENE GLYCOL | | Authors: | Zhang, Y.Y, Dong, S, Feng, Y.G, Chang, Y.G. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural characterization on a beta-agarase Aga86A_Wa from Wenyingzhuangia aestuarii reveals the prevalent methyl-galactose accommodation capacity of GH86 enzymes at subsite -1.
Carbohydr Polym, 306, 2023
|
|
8H56
 
 | | Crystal structure of Rep' of porcine circovirus type 2 | | Descriptor: | Isoform Rep' of Replication-associated protein | | Authors: | Guan, S.Y, Song, Y.F. | | Deposit date: | 2022-10-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of the dimerized of porcine circovirus type II replication-related protein Rep'.
Proteins, 91, 2023
|
|
8IVB
 
 | | K113-Ubiquitinated BAK | | Descriptor: | Bcl-2 homologous antagonist/killer, Ubiquitin | | Authors: | Dong, X, Cheng, P, Hou, Y.Z, Chen, Y.K, Liu, Z. | | Deposit date: | 2023-03-26 | | Release date: | 2024-01-31 | | Method: | SOLUTION NMR | | Cite: | Parkin-mediated ubiquitination inhibits BAK apoptotic activity by blocking its canonical hydrophobic groove.
Commun Biol, 6, 2023
|
|
2HU7
 
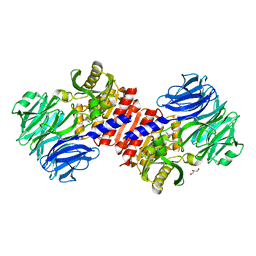 | | Binding of inhibitors by Acylaminoacyl peptidase | | Descriptor: | ACETYL GROUP, Acylamino-acid-releasing enzyme, GLYCEROL, ... | | Authors: | Kiss, A.L, Hornung, B, Radi, K, Gengeliczki, Z, Sztaray, B, Harmat, V, Polgar, L. | | Deposit date: | 2006-07-26 | | Release date: | 2007-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Acylaminoacyl Peptidase from Aeropyrum pernix K1 Thought to Be an Exopeptidase Displays Endopeptidase Activity
J.Mol.Biol., 368, 2007
|
|
2HU8
 
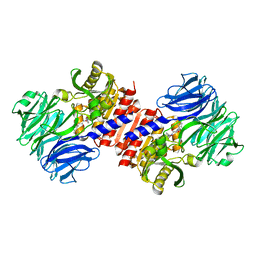 | | Binding of inhibitors by Acylaminoacyl peptidase | | Descriptor: | 2-AMINOBENZOIC ACID, Acylamino-acid-releasing enzyme, GLYCINE, ... | | Authors: | Kiss, A.L, Hornung, B, Radi, K, Gengeliczki, Z, Sztaray, B, Harmat, V, Polgar, L. | | Deposit date: | 2006-07-26 | | Release date: | 2007-05-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Acylaminoacyl Peptidase from Aeropyrum pernix K1 Thought to Be an Exopeptidase Displays Endopeptidase Activity
J.Mol.Biol., 368, 2007
|
|
7XUR
 
 | |
