7FI6
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | 分子名称: | MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | 著者 | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | 登録日 | 2021-07-30 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7FI5
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | 分子名称: | GLYCEROL, MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | 著者 | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | 登録日 | 2021-07-30 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7FI8
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | 分子名称: | MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | 著者 | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | 登録日 | 2021-07-30 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7FI7
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | 分子名称: | MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | 著者 | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | 登録日 | 2021-07-30 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7FI9
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | 分子名称: | GLYCEROL, MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | 著者 | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | 登録日 | 2021-07-30 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
5ZTE
 
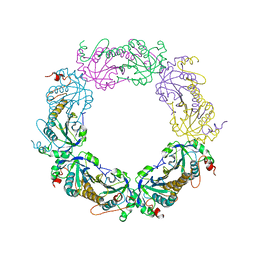 | | Crystal structure of PrxA C119S mutant from Arabidopsis thaliana | | 分子名称: | 2-Cys peroxiredoxin BAS1, chloroplastic | | 著者 | Yang, Y, Cai, W, Wang, J, Pan, W, Liu, L, Wang, M, Zhang, M. | | 登録日 | 2018-05-03 | | 公開日 | 2018-10-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of Arabidopsis thaliana peroxiredoxin A C119S mutant.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5YP6
 
 | | RORgamma (263-509) complexed with SRC2 and Compound 6 | | 分子名称: | N-[3'-cyano-4'-(2-methylpropyl)-2-(trifluoromethyl)biphenyl-4-yl]-2-[4-(ethylsulfonyl)phenyl]acetamide, Nuclear receptor ROR-gamma, SRC2 | | 著者 | Gao, M, Cai, W. | | 登録日 | 2017-11-01 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | From ROR gamma t Agonist to Two Types of ROR gamma t Inverse Agonists
ACS Med Chem Lett, 9, 2018
|
|
5YP5
 
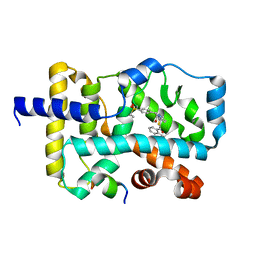 | | Crystal structure of RORgamma complexed with SRC2 and compound 5d | | 分子名称: | 2-[4-(ethylsulfonyl)phenyl]-N-{5-[2-(2-methylpropyl)benzoyl]-4-phenyl-1,3-thiazol-2-yl}acetamide, Nuclear receptor ROR-gamma, SRC2-2 peptide | | 著者 | Gao, M, Cai, W, Chunwa, C. | | 登録日 | 2017-11-01 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | From ROR gamma t Agonist to Two Types of ROR gamma t Inverse Agonists
ACS Med Chem Lett, 9, 2018
|
|
6N04
 
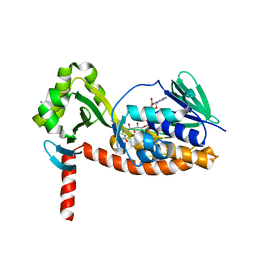 | | The X-ray crystal structure of AbsH3, an FAD dependent reductase from the Abyssomicin biosynthesis pathway in Streptomyces | | 分子名称: | AbsH3, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Clinger, J.A, Wang, X, Cai, W, Miller, M.D, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2018-11-06 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.998 Å) | | 主引用文献 | The crystal structure of AbsH3: A putative flavin adenine dinucleotide-dependent reductase in the abyssomicin biosynthesis pathway.
Proteins, 2020
|
|
5U1M
 
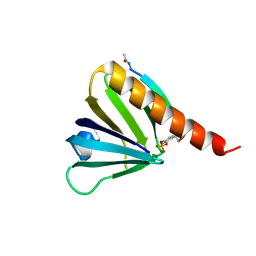 | |
6XO3
 
 | |
6XOJ
 
 | |
6XN6
 
 | |
6XPA
 
 | |
6Y1Z
 
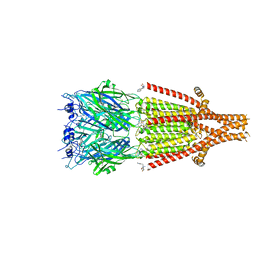 | | Mouse serotonin 5HT3 receptor in complex with palonosetron | | 分子名称: | (3~{a}~{S})-2-[(3~{S})-1-azabicyclo[2.2.2]octan-3-yl]-3~{a},4,5,6-tetrahydro-3~{H}-benzo[de]isoquinolin-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zarkadas, E, Perot, J, Nury, H. | | 登録日 | 2020-02-14 | | 公開日 | 2020-03-04 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (2.82 Å) | | 主引用文献 | The Binding of Palonosetron and Other Antiemetic Drugs to the Serotonin 5-HT3 Receptor.
Structure, 28, 2020
|
|
6DCH
 
 | |
8X84
 
 | |
8X83
 
 | |
8X82
 
 | |
8WGV
 
 | | BA.2(S375) Spike (S6P)/hACE2 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Wei, X, Zhang, Z. | | 登録日 | 2023-09-22 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
8WGW
 
 | | Local refinement of RBD-ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Wei, X, Zhang, Z. | | 登録日 | 2023-09-22 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
1ZZL
 
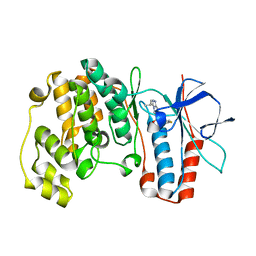 | | Crystal structure of P38 with triazolopyridine | | 分子名称: | 6-[4-(4-FLUOROPHENYL)-1,3-OXAZOL-5-YL]-3-ISOPROPYL[1,2,4]TRIAZOLO[4,3-A]PYRIDINE, Mitogen-activated protein kinase 14 | | 著者 | McClure, K.F, Han, S. | | 登録日 | 2005-06-14 | | 公開日 | 2005-09-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Theoretical and Experimental Design of Atypical Kinase Inhibitors: Application to p38 MAP Kinase.
J.Med.Chem., 48, 2005
|
|
6LYX
 
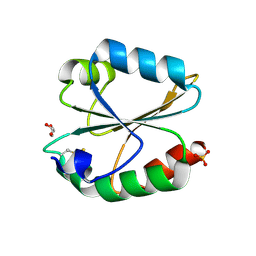 | | Crystal structure of oxidized ACHT1 | | 分子名称: | GLYCEROL, SULFATE ION, Thioredoxin-like 2-1, ... | | 著者 | Wang, J.C, Pan, W.M, Cai, W.G, Wang, M.Z, Zhang, M. | | 登録日 | 2020-02-16 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.696 Å) | | 主引用文献 | Structural insight into the biological functions of Arabidopsis thaliana ACHT1.
Int.J.Biol.Macromol., 158, 2020
|
|
4NIE
 
 | |
8DRP
 
 | |
