5Z0R
 
 | | Structural insight into the Zika virus capsid encapsulating the viral genome | | Descriptor: | Extracellular solute-binding protein family 1,viral genome protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, T, Zhao, Q, Yang, X, Chen, C, Yang, K, Wu, C, Zhang, T, Duan, Y, Xue, X, Mi, K, Ji, X, Wang, Z, Yang, H. | | Deposit date: | 2017-12-20 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insight into the Zika virus capsid encapsulating the viral genome.
Cell Res., 28, 2018
|
|
3LD1
 
 | | Crystal Structure of IBV Nsp2a | | Descriptor: | Replicase polyprotein 1a | | Authors: | Xu, Y, Cong, L, Wei, L, Fu, J, Chen, C, Yang, A, Tang, H, Bartlam, M, Rao, Z. | | Deposit date: | 2010-01-12 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | IBV nsp2 is an endosome-associated protein and viral pathogenicity factor
To be Published
|
|
3EWP
 
 | | complex of substrate ADP-ribose with IBV Nsp3 ADRP domain | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Xu, Y, Cong, L, Chen, C, Wei, L, Zhao, Q, Xu, X, Ma, Y, Bartlam, M, Rao, Z. | | Deposit date: | 2008-10-16 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of two coronavirus ADP-ribose-1''-monophosphatases and their complexes with ADP-Ribose: a systematic structural analysis of the viral ADRP domain.
J.Virol., 83, 2009
|
|
3EWQ
 
 | | HCov-229E Nsp3 ADRP domain | | Descriptor: | Non-structural protein 3 | | Authors: | Xu, Y, Cong, L, Chen, C, Wei, L, Zhao, Q, Xu, X, Ma, Y, Bartlam, M, Rao, Z. | | Deposit date: | 2008-10-16 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of two coronavirus ADP-ribose-1''-monophosphatases and their complexes with ADP-Ribose: a systematic structural analysis of the viral ADRP domain.
J.Virol., 83, 2009
|
|
3EWR
 
 | | complex of substrate ADP-ribose with HCoV-229E Nsp3 ADRP domain | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Xu, Y, Cong, L, Chen, C, Wei, L, Zhao, Q, Xu, X, Ma, Y, Bartlam, M, Rao, Z. | | Deposit date: | 2008-10-16 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of two coronavirus ADP-ribose-1''-monophosphatases and their complexes with ADP-Ribose: a systematic structural analysis of the viral ADRP domain.
J.Virol., 83, 2009
|
|
3EWO
 
 | | IBV Nsp3 ADRP domain | | Descriptor: | Non-structural protein 3 | | Authors: | Xu, Y, Cong, L, Chen, C, Wei, L, Zhao, Q, Xu, X, Ma, Y, Bartlam, M, Rao, Z. | | Deposit date: | 2008-10-16 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of two coronavirus ADP-ribose-1''-monophosphatases and their complexes with ADP-Ribose: a systematic structural analysis of the viral ADRP domain.
J.Virol., 83, 2009
|
|
3D23
 
 | | Main protease of HCoV-HKU1 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Zhao, Q, Chen, C, Li, S, Zou, Y. | | Deposit date: | 2008-05-07 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1.
J.Virol., 82, 2008
|
|
3FUQ
 
 | | Glycosylated SV2 and Gangliosides as Dual Receptors for Botulinum Neurotoxin Serotype F | | Descriptor: | BoNT/F (Neurotoxin type F) | | Authors: | Fu, Z, Chen, C, Barbieri, J.T, Kim, J.-J.P, Baldwin, M.R. | | Deposit date: | 2009-01-14 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Glycosylated SV2 and gangliosides as dual receptors for botulinum neurotoxin serotype F
Biochemistry, 48, 2009
|
|
3FUO
 
 | | The Crystal structure of receptor binding domain of botulinum neurotoxin serotype A | | Descriptor: | Botulinum neurotoxin type A | | Authors: | Fu, Z, Chen, C, Barbieri, J.T, Kim, J.-J.P, Baldwin, M.R. | | Deposit date: | 2009-01-14 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glycosylated SV2 and gangliosides as dual receptors for botulinum neurotoxin serotype F
Biochemistry, 48, 2009
|
|
5Y6N
 
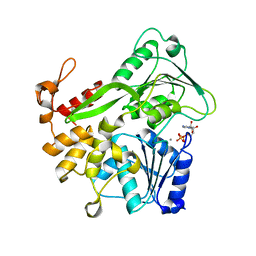 | | Zika virus helicase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Helicase domain from Genome polyprotein, MANGANESE (II) ION | | Authors: | Yang, X.Y, Chen, C, Tian, H.L, Chi, H, Mu, Z.Y, Zhang, T.Q, Yang, K.L, Zhao, Q, Liu, X.H, Wang, Z.F, Ji, X.Y, Yang, H.T. | | Deposit date: | 2017-08-12 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Mechanism of ATP hydrolysis by the Zika virus helicase.
FASEB J., 32, 2018
|
|
5Y6M
 
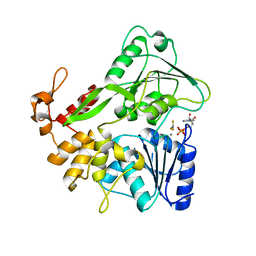 | | Zika virus helicase in complex with ADP-AlF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase domain from Genome polyprotein, ... | | Authors: | Yang, X.Y, Chen, C, Tian, H.L, Chi, H, Mu, Z.Y, Zhang, T.Q, Yang, K.L, Zhao, Q, Liu, X.H, Wang, Z.F, Ji, X.Y, Yang, H.T. | | Deposit date: | 2017-08-12 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Mechanism of ATP hydrolysis by the Zika virus helicase.
FASEB J., 32, 2018
|
|
5Z0V
 
 | | Structural insight into the Zika virus capsid encapsulating the viral genome | | Descriptor: | Extracellular solute-binding protein family 1,viral genome protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, T, Zhao, Q, Yang, X, Chen, C, Yang, K, Wu, C, Zhang, T, Duan, Y, Xue, X, Mi, K, Ji, X, Wang, Z, Yang, H. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Structural insight into the Zika virus capsid encapsulating the viral genome.
Cell Res., 28, 2018
|
|
6KZ6
 
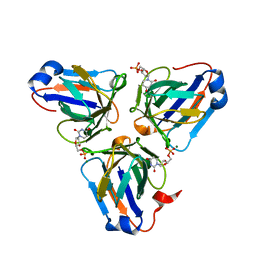 | | Crystal structure of ASFV dUTPase | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R, MAGNESIUM ION | | Authors: | Guo, Y, Chen, C, Li, G.B, Cao, L, Wang, C.W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Structural Insight into African Swine Fever Virus dUTPase Reveals a Novel Folding Pattern in the dUTPase Family.
J.Virol., 94, 2020
|
|
6JK2
 
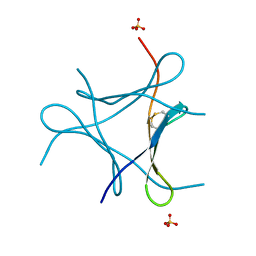 | | Crystal structure of a mini fungal lectin, PhoSL | | Descriptor: | Lectin, SULFATE ION | | Authors: | Lou, Y.C, Chou, C.C, Yeh, H.H, Chien, C.Y, Sushant, S, Chen, C, Hsu, C.H. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structural insights into the role of N-terminal integrity in PhoSL for core-fucosylated N-glycan recognition.
Int.J.Biol.Macromol., 255, 2023
|
|
6IDO
 
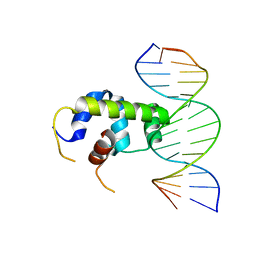 | | Crystal structure of Klebsiella pneumoniae sigma4 of sigmaS fusing with the RNA polymerase beta-flap-tip-helix in complex with -35 element DNA | | Descriptor: | DNA (5'-D(P*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*AP*AP*TP*CP*G)-3'), DNA (5'-D(P*GP*AP*TP*TP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*C)-3'), RNA polymerase sigma factor RpoS,RNA polymerase beta-flap-tip-helix | | Authors: | Lou, Y.C, Chien, C.Y, Chen, C, Hsu, C.H. | | Deposit date: | 2018-09-10 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.748 Å) | | Cite: | Structural basis for -35 element recognition by sigma4chimera proteins and their interactions with PmrA response regulator.
Proteins, 88, 2020
|
|
6JK3
 
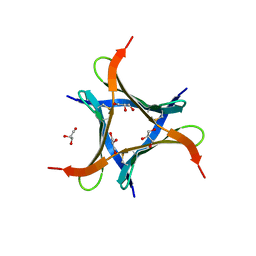 | | Crystal structure of a mini fungal lectin, PhoSL in complex with core-fucosylated chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Lectin | | Authors: | Lou, Y.C, Chou, C.C, Yeh, H.H, Chien, C.Y, Sushant, S, Chen, C, Hsu, C.H. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural insights into the role of N-terminal integrity in PhoSL for core-fucosylated N-glycan recognition.
Int.J.Biol.Macromol., 255, 2023
|
|
7DNU
 
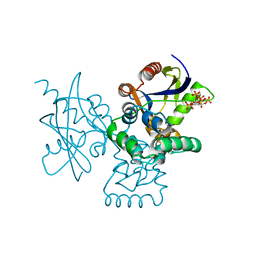 | | mRNA-decapping enzyme g5Rp with inhibitor insp6 complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, mRNA-decapping protein g5R | | Authors: | Yang, Y, Chen, C, Li, L, Li, X.H, Su, D. | | Deposit date: | 2020-12-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Structural Insight into Molecular Inhibitory Mechanism of InsP 6 on African Swine Fever Virus mRNA-Decapping Enzyme g5Rp.
J.Virol., 96, 2022
|
|
7DNT
 
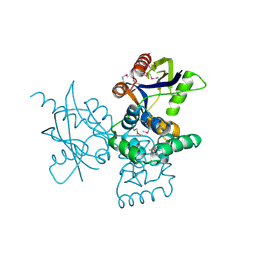 | | mRNA-decapping enzyme g5Rp | | Descriptor: | mRNA-decapping protein g5R | | Authors: | Yang, Y, Chen, C, Li, L, Li, X.H, Su, D. | | Deposit date: | 2020-12-10 | | Release date: | 2022-03-09 | | Last modified: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insight into Molecular Inhibitory Mechanism of InsP 6 on African Swine Fever Virus mRNA-Decapping Enzyme g5Rp.
J.Virol., 96, 2022
|
|
6LJ9
 
 | |
6LJB
 
 | |
5X3L
 
 | |
5YB9
 
 | | Crystal structure of a dimeric cyclophilin A from T.vaginalis | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Cho, C.C, Lin, M.H, Chou, C.C, Martin, T, Chen, C, Hsu, C.H. | | Deposit date: | 2017-09-04 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.276 Å) | | Cite: | Structural basis of interaction between dimeric cyclophilin 1 and Myb1 transcription factor in Trichomonas vaginalis
Sci Rep, 8, 2018
|
|
5XZO
 
 | | Crystal structure of GH10 xylanase XYL10C from Bispora. sp MEY-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-xylanase | | Authors: | You, S, Chen, C, Tu, T, Guo, R.T, Luo, H, Yao, B. | | Deposit date: | 2017-07-13 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of GH10 xylanase XYL10C from Bispora. sp MEY-1
To Be Published
|
|
5YBA
 
 | | Dimeric Cyclophilin from T.vaginalis in complex with Myb1 peptide | | Descriptor: | Myb1 peptide, Peptidyl-prolyl cis-trans isomerase | | Authors: | Cho, C.C, Lin, M.H, Martin, T, Chou, C.C, Chen, C, Hsu, C.H. | | Deposit date: | 2017-09-04 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.062 Å) | | Cite: | Structural basis of interaction between dimeric cyclophilin 1 and Myb1 transcription factor in Trichomonas vaginalis
Sci Rep, 8, 2018
|
|
8J3O
 
 | | Formate dehydrogenase wild-type enzyme from Candida dubliniensis complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Formate dehydrogenase, MAGNESIUM ION | | Authors: | Ma, W, Zheng, Y.C, Geng, Q, Chen, C. | | Deposit date: | 2023-04-17 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineering a Formate Dehydrogenase for NADPH Regeneration.
Chembiochem, 24, 2023
|
|
