7ADW
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2,4'-Dimethylpropiophenone. | | Descriptor: | 2-methyl-1-(4-methylphenyl)propan-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7A1U
 
 | | Structure of SARS-CoV-2 Main Protease bound to Fusidic Acid. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, FUSIDIC ACID, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6Q6Q
 
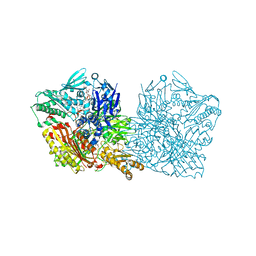 | | Human aldehyde oxidase SNP G1269R | | Descriptor: | Aldehyde oxidase, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mota, C, Coelho, C, Santos-Silva, T, Romao, M.J. | | Deposit date: | 2018-12-11 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.10003781 Å) | | Cite: | Human aldehyde oxidase (hAOX1): structure determination of the Moco-free form of the natural variant G1269R and biophysical studies of single nucleotide polymorphisms.
Febs Open Bio, 9, 2019
|
|
6WEB
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
6WEC
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
8TFO
 
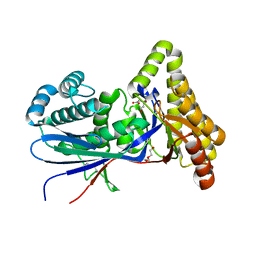 | | Structure of MKvar | | Descriptor: | (R)-MEVALONATE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peat, T.S, Newman, J, Esquirol, L, Nebl, T, Scott, C, Vickers, C, Sainsbury, F. | | Deposit date: | 2023-07-11 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of novel mevalonate kinases from the tardigrade Ramazzottius varieornatus and the psychrophilic archaeon Methanococcoides burtonii.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8TEB
 
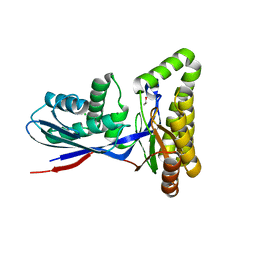 | | Structure of MKbur | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Peat, T.S, Newman, J, Esquirol, L, Nebl, T, Scott, C, Vickers, C, Sainsbury, F. | | Deposit date: | 2023-07-06 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of novel mevalonate kinases from the tardigrade Ramazzottius varieornatus and the psychrophilic archaeon Methanococcoides burtonii.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8TXA
 
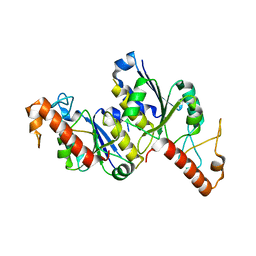 | | Apo Structure of (N1G37) tRNA Methyltransferase from Mycobacterium marinum | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Balsamo, A, Bruno, C, Edele, C, Fabian, T, Jannotta, R, Lee, R, Schryver, D, Warsaw, J, Warsaw, L, Stojanoff, V, Battaile, K, Perez, A, Bolen, R. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Apo Structure of (N1G37) tRNA Methyltransferase from Mycobacterium marinum
To Be Published
|
|
966C
 
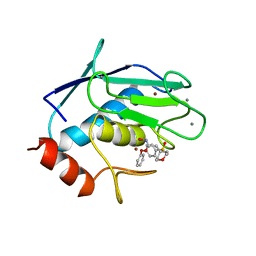 | | CRYSTAL STRUCTURE OF FIBROBLAST COLLAGENASE-1 COMPLEXED TO A DIPHENYL-ETHER SULPHONE BASED HYDROXAMIC ACID | | Descriptor: | CALCIUM ION, MMP-1, N-HYDROXY-2-[4-(4-PHENOXY-BENZENESULFONYL)-TETRAHYDRO-PYRAN-4-YL]-ACETAMIDE, ... | | Authors: | Lovejoy, B, Welch, A, Carr, S, Luong, C, Broka, C, Hendricks, R.T, Campbell, J, Walker, K, Martin, R, Van Wart, H, Browner, M.F. | | Deposit date: | 1998-08-07 | | Release date: | 1999-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors.
Nat.Struct.Biol., 6, 1999
|
|
8WZD
 
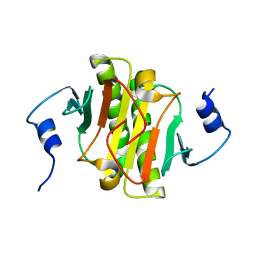 | |
8X2Q
 
 | | The Crystal Structure of APC from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Adenomatous polyposis coli protein | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Bao, C. | | Deposit date: | 2023-11-10 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of APC from Biortus.
To Be Published
|
|
8X5Z
 
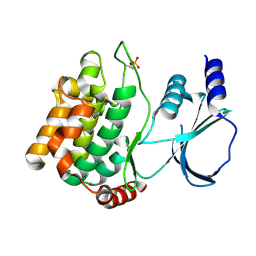 | |
8XQU
 
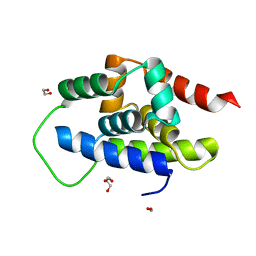 | | The Crystal Structure of ClpC1-NTD from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent Clp protease ATP-binding subunit ClpC1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Ni, C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of ClpC1-NTD from Biortus.
To Be Published
|
|
8Y9A
 
 | |
8XQK
 
 | | The Crystal Structure of Apaf from Biortus. | | Descriptor: | Apoptotic protease-activating factor 1, PHOSPHATE ION | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Ni, C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Crystal Structure of Apaf from Biortus.
To Be Published
|
|
8X7A
 
 | | Treprostinil bound Prostacyclin Receptor G protein complex | | Descriptor: | 2-[[(1~{R},2~{R},3~{a}~{S},9~{a}~{S})-2-oxidanyl-1-[(3~{S})-3-oxidanyloctyl]-2,3,3~{a},4,9,9~{a}-hexahydro-1~{H}-cyclopenta[g]naphthalen-5-yl]oxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.J, Jin, S, Zhang, H, Xu, Y, Hu, W, Jiang, Y, Chen, C, Wang, D.W, Xu, H.E, Wu, C. | | Deposit date: | 2023-11-23 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Molecular recognition and activation of the prostacyclin receptor by anti-pulmonary arterial hypertension drugs.
Sci Adv, 10, 2024
|
|
8X79
 
 | | MRE-269 bound Prostacyclin Receptor G protein complex | | Descriptor: | 2-[4-[(5,6-diphenylpyrazin-2-yl)-propan-2-yl-amino]butoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.J, Jin, S, Zhang, H, Xu, Y, Hu, W, Jiang, Y, Chen, C, Wang, D.W, Xu, H.E, Wu, C. | | Deposit date: | 2023-11-23 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Molecular recognition and activation of the prostacyclin receptor by anti-pulmonary arterial hypertension drugs.
Sci Adv, 10, 2024
|
|
8XQD
 
 | |
8XI8
 
 | |
8XX1
 
 | |
8X39
 
 | |
8X3A
 
 | |
8SNX
 
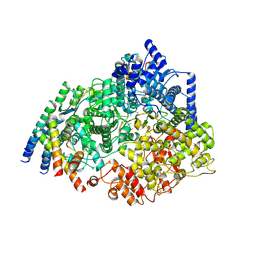 | | Cryo-EM structure of the respiratory syncytial virus polymerase (L:P) bound to the leader promoter | | Descriptor: | Phosphoprotein, RNA (5'-R(*UP*UP*UP*UP*UP*CP*GP*CP*GP*U)-3'), RNA-directed RNA polymerase L | | Authors: | Cao, D, Gao, Y, Chen, Z, Gooneratne, I, Roesler, C, Mera, C, Liang, B. | | Deposit date: | 2023-04-28 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of the promoter-bound respiratory syncytial virus polymerase.
Nature, 625, 2024
|
|
8SNY
 
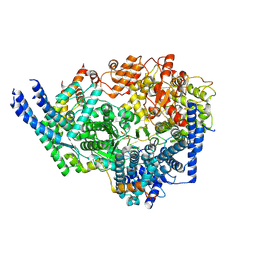 | | Cryo-EM structure of the respiratory syncytial virus polymerase (L:P) bound to the trailer complementary promoter | | Descriptor: | Phosphoprotein, RNA (5'-R(*UP*UP*UP*UP*UP*CP*UP*CP*GP*U)-3'), RNA-directed RNA polymerase L | | Authors: | Cao, D, Gao, Y, Chen, Z, Gooneratne, I, Roesler, C, Mera, C, Liang, B. | | Deposit date: | 2023-04-28 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structures of the promoter-bound respiratory syncytial virus polymerase.
Nature, 625, 2024
|
|
8T8D
 
 | | Structure of Helicobacter pylori adhesin A, HpaA | | Descriptor: | ACETATE ION, GLYCEROL, Neuraminyllactose-binding hemagglutinin | | Authors: | Martini, C, Calmettes, C. | | Deposit date: | 2023-06-22 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Unraveling the crystal structure of the HpaA adhesin: insights into cell adhesion function and epitope localization of a Helicobacter pylori vaccine candidate.
Mbio, 15, 2024
|
|
