5MLD
 
 | | Crystal Structure of RosB with bound intermediate AFP (8-demethyl-8-aminoriboflavin-5'-phosphate) | | Descriptor: | 8-demethyl-8-aminoriboflavin-5'-phosphate, BRAMP domain protein | | Authors: | Konjik, V, Bruenle, S, Demmer, U, Vanselow, A, Sandhoff, R, Mack, M, Ermler, U. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of RosB gives insights into the reaction mechanism of this first known member of a new family of flavodoxin-like enzymes
To be published
|
|
6QZH
 
 | | Structure of the human CC Chemokine Receptor 7 in complex with the intracellular allosteric antagonist Cmp2105 and the insertion protein Sialidase NanA | | Descriptor: | 3-[[4-[[(1~{R})-2,2-dimethyl-1-(5-methylfuran-2-yl)propyl]amino]-1,1-bis(oxidanylidene)-1,2,5-thiadiazol-3-yl]amino]-~{N},~{N},6-trimethyl-2-oxidanyl-benzamide, C-C chemokine receptor type 7,Sialidase A,C-C chemokine receptor type 7, D(-)-TARTARIC ACID, ... | | Authors: | Jaeger, K, Bruenle, S, Weinert, T, Guba, W, Muehle, J, Miyazaki, T, Weber, M, Furrer, A, Haenggi, N, Tetaz, T, Huang, C.Y, Mattle, D, Vonach, J.M, Gast, A, Kuglstatter, A, Rudolph, M.G, Nogly, P, Benz, J, Dawson, R.J.P, Standfuss, J. | | Deposit date: | 2019-03-11 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Allosteric Ligand Recognition in the Human CC Chemokine Receptor 7.
Cell, 178, 2019
|
|
6RKD
 
 | | Molybdenum storage protein under turnover conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MO(VI)(=O)(OH)2 CLUSTER, ... | | Authors: | Bruenle, S, Mills, D.J, Vonck, J, Ermler, U. | | Deposit date: | 2019-04-30 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molybdate pumping into the molybdenum storage protein via an ATP-powered piercing mechanism.
Proc.Natl.Acad.Sci.USA, 2019
|
|
9FKA
 
 | | Cryo-EM structure of the reduced cytochrome bd oxidase from M. tuberculosis | | Descriptor: | (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, CARDIOLIPIN, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, ... | | Authors: | Kayastha, K, Bruenle, S. | | Deposit date: | 2024-06-03 | | Release date: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Menaquinone-specific turnover by Mycobacterium tuberculosis cytochrome bd is redox regulated by the Q-loop disulfide bond.
J.Biol.Chem., 301, 2024
|
|
8A6E
 
 | | 100 picosecond light activated crystal structure of bovine rhodopsin in Lipidic Cubic Phase (SACLA) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C.J, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P.J.M, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C.M, Diethelm, A.D, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-06-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
8A6D
 
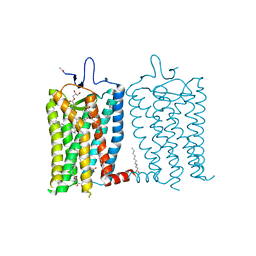 | | 10 picosecond light activated crystal structure of bovine rhodopsin in Lipidic Cubic Phase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C.J, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P.J.M, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C.M, Diethelm, A.D, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-06-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
8A6C
 
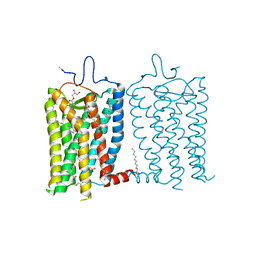 | | 1 picosecond light activated crystal structure of bovine rhodopsin in Lipidic Cubic Phase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C.J, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P.J.M, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C.M, Diethelm, A.D, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-06-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
5MJI
 
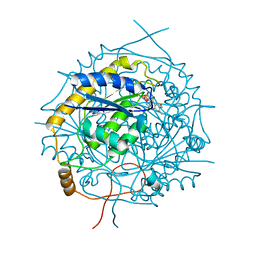 | | Crystal Structure of RosB with bound intermediate OHC-RP (8-demethyl-8-formylriboflavin-5'-phosphate) | | Descriptor: | BRAMP domain protein, [(2~{S},3~{R},4~{R})-5-[8-methanoyl-7-methyl-2,4-bis(oxidanylidene)-1~{H}-benzo[g]pteridin-10-ium-10-yl]-2,3,4-tris(oxidanyl)pentyl] dihydrogen phosphate | | Authors: | Konjik, V, Bruenle, S, Demmer, U, Vanselow, A, Sandhoff, R, Mack, M, Ermler, U. | | Deposit date: | 2016-12-01 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of RosB: Insights into the Reaction Mechanism of the First Member of a Family of Flavodoxin-like Enzymes.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7ZBE
 
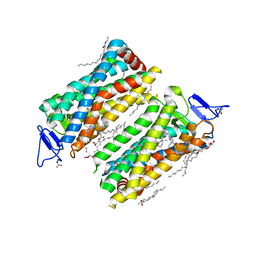 | | Dark state crystal structure of bovine rhodopsin in Lipidic Cubic Phase (SwissFEL) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C, Diethelm, A, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-03-23 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
7ZBC
 
 | | Dark state crystal structure of bovine rhodopsin in Lipidic Cubic Phase (SACLA) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C, Diethelm, A, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, S, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-03-23 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
9GO1
 
 | | C1C2 Channelrhodopsin - SMX Dark structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Archaeal-type opsin 1,Archaeal-type opsin 2, RETINAL | | Authors: | Mulder, M, Weinert, T, Skopintsev, P, Bruenle, S, Broser, M, Hegemann, P, Standfuss, J. | | Deposit date: | 2024-09-04 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Insights Into the Opening Mechanism of C1C2 Channelrhodopsin.
J.Am.Chem.Soc., 147, 2025
|
|
9GO2
 
 | | C1C2 Channelrhodopsin - SMX Light activated structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Archaeal-type opsin 1,Archaeal-type opsin 2, RETINAL | | Authors: | Mulder, M, Weinert, T, Skopintsev, P, Bruenle, S, Broser, M, Hegemann, P, Standfuss, J. | | Deposit date: | 2024-09-04 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights Into the Opening Mechanism of C1C2 Channelrhodopsin.
J.Am.Chem.Soc., 147, 2025
|
|
8RII
 
 | |
8RG2
 
 | |
8RFZ
 
 | |
8BTW
 
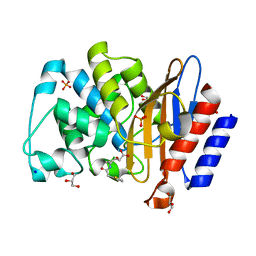 | | Structure of D179N BlaC from Mycobacterium tuberculosis in complex with vaborbactam | | Descriptor: | Beta-lactamase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Chikunova, A, Bruenle, S, Ubbink, M. | | Deposit date: | 2022-12-02 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Asp179 in the class A beta-lactamase from Mycobacterium tuberculosis is a conserved yet not essential residue due to epistasis.
Febs J., 290, 2023
|
|
8BTV
 
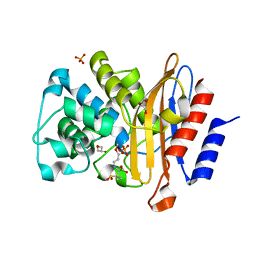 | | Structure of D179N BlaC from Mycobacterium tuberculosis bound to the trans-enamine adduct of sulbactam | | Descriptor: | Beta-lactamase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Chikunova, A, Bruenle, S, Ubbink, M. | | Deposit date: | 2022-12-02 | | Release date: | 2023-07-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Asp179 in the class A beta-lactamase from Mycobacterium tuberculosis is a conserved yet not essential residue due to epistasis.
Febs J., 290, 2023
|
|
8BV4
 
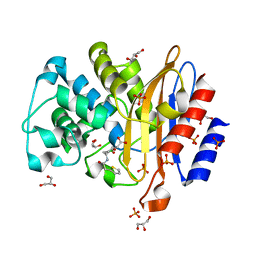 | | Structure of BlaC from Mycobacterium tuberculosis in complex with vaborbactam | | Descriptor: | Beta-lactamase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Chikunova, A, Bruenle, S, Ubbink, M. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Asp179 in the class A beta-lactamase from Mycobacterium tuberculosis is a conserved yet not essential residue due to epistasis.
Febs J., 290, 2023
|
|
8BTU
 
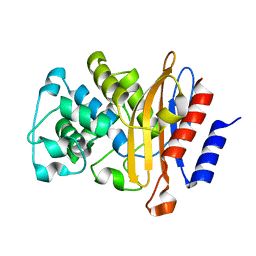 | |
6RQP
 
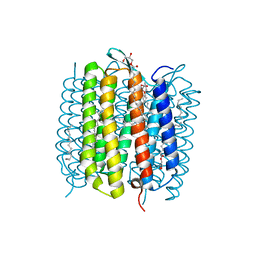 | | Steady-state-SMX dark state structure of bacteriorhodopsin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Weinert, T, Skopintsev, P, James, D, Kekilli, D, Furrer, A, Bruenle, S, Mous, S, Nogly, P, Standfuss, J. | | Deposit date: | 2019-05-16 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography.
Science, 365, 2019
|
|
6RQO
 
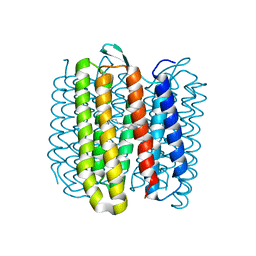 | | Steady-state-SMX activated state structure of bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Weinert, T, Skopintsev, P, James, D, Kekilli, D, Furrer, A, Bruenle, S, Mous, S, Nogly, P, Standfuss, J. | | Deposit date: | 2019-05-16 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography.
Science, 365, 2019
|
|
6RNJ
 
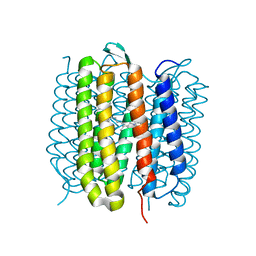 | | TR-SMX closed state structure (0-5ms) of bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Weinert, T, Skopintsev, P, James, D, Kekilli, D, Furrer, F, Bruenle, S, Mous, S, Nogly, P, Standfuss, J. | | Deposit date: | 2019-05-08 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography.
Science, 365, 2019
|
|
6RKE
 
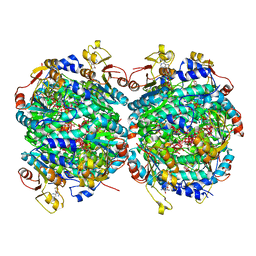 | | Molybdenum storage protein - P212121, ADP, molybdate | | Descriptor: | (mu3-oxo)-tris(mu2-oxo)-nonakisoxo-trimolybdenum (VI), ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ermler, U, Bruenle, S. | | Deposit date: | 2019-04-30 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molybdate pumping into the molybdenum storage protein via an ATP-powered piercing mechanism.
Proc.Natl.Acad.Sci.USA, 2019
|
|
6TK6
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: Dark structure in neutral conditions with attached light datasets at 800fs, 2ps, 100ps, 1ns, 16ns, 1us, 30us, 150us, 1ms and 20ms | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
6TK4
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: 1ns+16ns structure of KR2 with extrapolated, light and dark datasets | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
