4WFH
 
 | |
4WFG
 
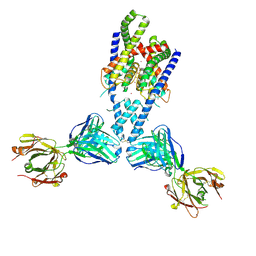
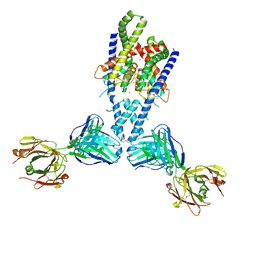 | | Human TRAAK K+ channel in a Tl+ bound conductive conformation | | Descriptor: | ANTI-TRAAK ANTIBODY 13E9 FAB FRAGMENT HEAVY CHAIN, ANTI-TRAAK ANTIBODY 13E9 FAB FRAGMENT LIGHT CHAIN, CALCIUM ION, ... | | Authors: | Brohawn, S.G, MacKinnon, R. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Physical mechanism for gating and mechanosensitivity of the human TRAAK K+ channel.
Nature, 516, 2014
|
|
4WFE
 
 | | Human TRAAK K+ channel in a K+ bound conductive conformation | | Descriptor: | ANTI-TRAAK ANTIBODY 13E9 FAB FRAGMENT HEAVY CHAIN, ANTI-TRAAK ANTIBODY 13E9 FAB FRAGMENT LIGHT CHAIN, CALCIUM ION, ... | | Authors: | Brohawn, S.G, MacKinnon, R. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Physical mechanism for gating and mechanosensitivity of the human TRAAK K+ channel.
Nature, 516, 2014
|
|
4I9W
 
 | |
6PIS
 
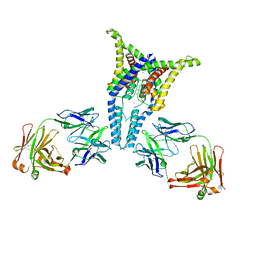 | |
4WFF
 
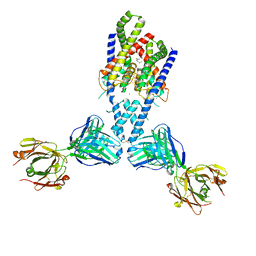 | |
3EWE
 
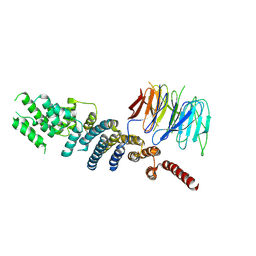 | | Crystal Structure of the Nup85/Seh1 Complex | | Descriptor: | Nucleoporin NUP85, Nucleoporin SEH1 | | Authors: | Brohawn, S.G, Leksa, N.C, Rajashankar, K.R, Schwartz, T.U. | | Deposit date: | 2008-10-14 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural evidence for common ancestry of the nuclear pore complex and vesicle coats.
Science, 322, 2008
|
|
3UM7
 
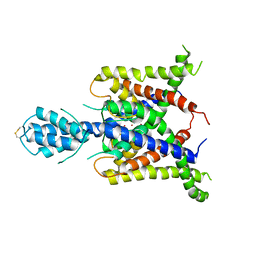 | |
3JRP
 
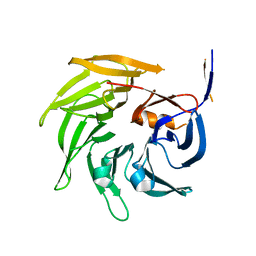 | | SEC13 with NUP145C (AA109-179) insertion blade | | Descriptor: | FUSION PROTEIN OF PROTEIN TRANSPORT PROTEIN SEC13 AND NUCLEOPORIN NUP145 | | Authors: | Brohawn, S.G, Gogola, M, Schwartz, T.U. | | Deposit date: | 2009-09-08 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3JRO
 
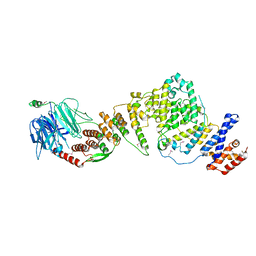 | | NUP84-NUP145C-SEC13 edge element of the NPC lattice | | Descriptor: | Fusion Protein of Protein Transport Protein SEC13 and Nucleoporin NUP145, Nucleoporin NUP84 | | Authors: | Brohawn, S.G, Schwartz, T.U. | | Deposit date: | 2009-09-08 | | Release date: | 2009-10-27 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (4.004 Å) | | Cite: | Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6XDC
 
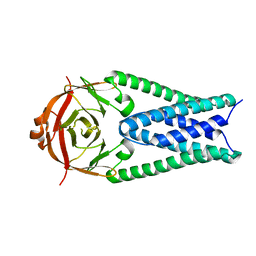 | |
7SK0
 
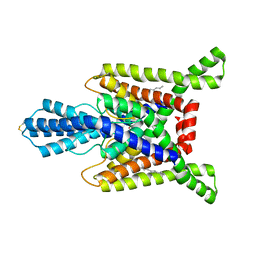 | | TWIK1 in MSP1D1 lipid nanodisc at pH 7.4 | | Descriptor: | N-OCTANE, POTASSIUM ION, Potassium channel subfamily K member 1 | | Authors: | Turney, T.S, Brohawn, S.G. | | Deposit date: | 2021-10-19 | | Release date: | 2021-11-24 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural Basis for pH-gating of the K + channel TWIK1 at the selectivity filter.
Nat Commun, 13, 2022
|
|
7SK1
 
 | | TWIK1 in MSP1E3D1 Lipid Nanodisc at pH 5.5 | | Descriptor: | N-OCTANE, POTASSIUM ION, Potassium channel subfamily K member 1, ... | | Authors: | Turney, T.S, Brohawn, S.G. | | Deposit date: | 2021-10-19 | | Release date: | 2021-11-24 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Structural Basis for pH-gating of the K + channel TWIK1 at the selectivity filter.
Nat Commun, 13, 2022
|
|
9C49
 
 | |
6UKN
 
 | | Cryo-EM structure of the potassium-chloride cotransporter KCC4 in lipid nanodiscs | | Descriptor: | CHLORIDE ION, POTASSIUM ION, Solute carrier family 12 member 7, ... | | Authors: | Reid, M.S, Kern, D.M, Brohawn, S.G. | | Deposit date: | 2019-10-05 | | Release date: | 2020-06-10 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Cryo-EM structure of the potassium-chloride cotransporter KCC4 in lipid nanodiscs.
Elife, 9, 2020
|
|
6WLV
 
 | | TASK2 in MSP1D1 lipid nanodisc at pH 6.5 | | Descriptor: | POTASSIUM ION, Potassium channel TASK2 | | Authors: | Li, B, Brohawn, S.G. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-15 | | Last modified: | 2020-10-28 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis for pH gating of the two-pore domain K + channel TASK2.
Nature, 586, 2020
|
|
6WM0
 
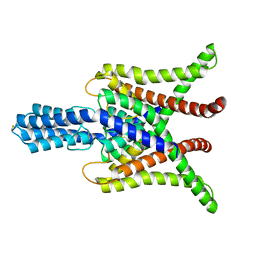 | | TASK2 in MSP1D1 lipid nanodisc at pH 8.5 | | Descriptor: | POTASSIUM ION, Potassium channel TASK2 | | Authors: | Li, B, Brohawn, S.G. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-15 | | Last modified: | 2020-10-28 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis for pH gating of the two-pore domain K + channel TASK2.
Nature, 586, 2020
|
|
3HXR
 
 | |
7RTT
 
 | | Cryo-EM structure of a TTYH2 cis-dimer | | Descriptor: | CALCIUM ION, Protein tweety homolog 2, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, B, Brohawn, S.G, Hoel, C.M. | | Deposit date: | 2021-08-15 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of tweety homolog proteins TTYH2 and TTYH3 reveal a Ca 2+ -dependent switch from intra- to intermembrane dimerization.
Nat Commun, 12, 2021
|
|
7RTU
 
 | | Cryo-EM structure of a TTYH2 trans-dimer | | Descriptor: | Protein tweety homolog 2, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, B, Brohawn, S.G, Hoel, C.M. | | Deposit date: | 2021-08-15 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Structures of tweety homolog proteins TTYH2 and TTYH3 reveal a Ca 2+ -dependent switch from intra- to intermembrane dimerization.
Nat Commun, 12, 2021
|
|
7RTW
 
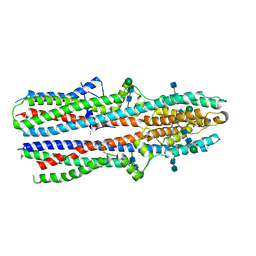 | | Cryo-EM structure of a TTYH3 cis-dimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, B, Brohawn, S.G, Hoel, C.M. | | Deposit date: | 2021-08-15 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structures of tweety homolog proteins TTYH2 and TTYH3 reveal a Ca 2+ -dependent switch from intra- to intermembrane dimerization.
Nat Commun, 12, 2021
|
|
7RTV
 
 | | Cryo-EM structure of monomeric TTYH2 | | Descriptor: | Protein tweety homolog 2, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, B, Brohawn, S.G, Hoel, C.M. | | Deposit date: | 2021-08-15 | | Release date: | 2021-09-22 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structures of tweety homolog proteins TTYH2 and TTYH3 reveal a Ca 2+ -dependent switch from intra- to intermembrane dimerization.
Nat Commun, 12, 2021
|
|
6NZZ
 
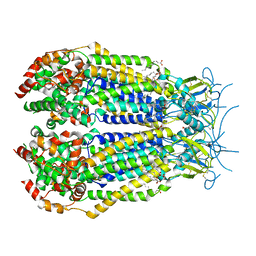 | | LRRC8A-DCPIB in MSP1E3D1 nanodisc expanded state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 4-{[(2S)-2-butyl-6,7-dichloro-2-cyclopentyl-1-oxo-2,3-dihydro-1H-inden-5-yl]oxy}butanoic acid, Volume-regulated anion channel subunit LRRC8A | | Authors: | Kern, D.M, Hite, R.K, Brohawn, S.G. | | Deposit date: | 2019-02-14 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM structures of the DCPIB-inhibited volume-regulated anion channel LRRC8A in lipid nanodiscs.
Elife, 8, 2019
|
|
6O00
 
 | |
6NZW
 
 | | LRRC8A-DCPIB in MSP1E3D1 nanodisc constricted state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 4-{[(2S)-2-butyl-6,7-dichloro-2-cyclopentyl-1-oxo-2,3-dihydro-1H-inden-5-yl]oxy}butanoic acid, Volume-regulated anion channel subunit LRRC8A | | Authors: | Kern, D.M, Hite, R.K, Brohawn, S.G. | | Deposit date: | 2019-02-14 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structures of the DCPIB-inhibited volume-regulated anion channel LRRC8A in lipid nanodiscs.
Elife, 8, 2019
|
|
