4RIE
 
 | | Landomycin Glycosyltransferase LanGT2 | | Descriptor: | Glycosyl transferase homolog | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RIF
 
 | | Landomycin Glycosyltransferase LanGT2, carbasugar substrate complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-{[(R)-{[(1S,3R,4R,5S)-3,4-dihydroxy-5-methylcyclohexyl]oxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-3,4-dihydrothymidine, Glycosyl transferase homolog | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RIG
 
 | | Chimeric Glycosyltransferase LanGT2S8Ac | | Descriptor: | Glycosyl transferase, MAGNESIUM ION | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RIH
 
 | | Chimeric Glycosyltransferase LanGT2S8Ac, carbasugar substrate complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-{[(R)-{[(1S,3R,4R,5S)-3,4-dihydroxy-5-methylcyclohexyl]oxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-3,4-dihydrothymidine, Glycosyl transferase homolog,Glycosyl transferase, MAGNESIUM ION | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RII
 
 | | Chimeric Glycosyltransferase LanGT2S8Ac, TDP complex | | Descriptor: | Glycosyl transferase homolog,Glycosyl transferase, MAGNESIUM ION, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4KAP
 
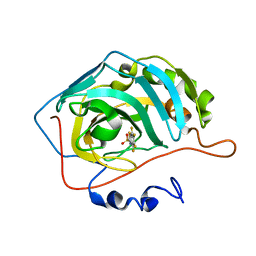 | | The Binding of Benzoarylsulfonamide Ligands to Human Carbonic Anhydrase is Insensitive to Formal Fluorination of the Ligand | | Descriptor: | 4,5,6,7-tetrafluoro-1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Lange, H, Lockett, M.R, Breiten, B, Heroux, A, Sherman, W, Rappoport, D, Yau, P.O, Snyder, P.W, Whitesides, G.M. | | Deposit date: | 2013-04-22 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Binding of Benzoarylsulfonamide Ligands to Human Carbonic Anhydrase is Insensitive to Formal Fluorination of the Ligand.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
7R5B
 
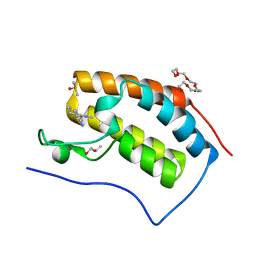 | | Crystal structure of BRD4(1) in complex with the inhibitor MPM2 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 1-(3-aminophenyl)-3-methyl-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrol-4-one, Bromodomain-containing protein 4, ... | | Authors: | Huegle, M. | | Deposit date: | 2022-02-10 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A novel pan-selective bromodomain inhibitor for epigenetic drug design
Eur.J.Med.Chem., 249, 2023
|
|
7AXR
 
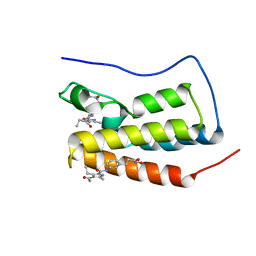 | | Crystal structure of BRD4(1) bound to the dual BET-HDAC inhibitor LSH24 | | Descriptor: | 4-acetyl-3-ethyl-N-(3-(3-(hydroxyamino)-3-oxopropyl)phenyl)-5-methyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Huegle, M. | | Deposit date: | 2020-11-10 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4-Acyl Pyrrole Capped HDAC Inhibitors: A New Scaffold for Hybrid Inhibitors of BET Proteins and Histone Deacetylases as Antileukemia Drug Leads.
J.Med.Chem., 64, 2021
|
|
7B1T
 
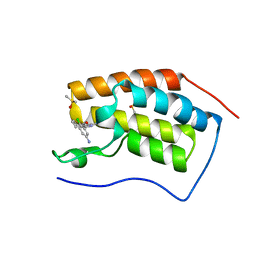 | | Crystal structure of BRD4(1) in complex with the inhibitor MPM6 | | Descriptor: | 3-(5-azanyl-2-chloranyl-phenyl)-1-methyl-4,7-dihydro-2~{H}-cyclohepta[c]pyrrol-8-one, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE | | Authors: | Huegle, M. | | Deposit date: | 2020-11-25 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A novel pan-selective bromodomain inhibitor for epigenetic drug design.
Eur.J.Med.Chem., 249, 2023
|
|
5NRW
 
 | |
5NU3
 
 | | Crystal structure of the human bromodomain of CREBBP bound to the inhibitor XDM-CBP | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CREB-binding protein, SULFATE ION, ... | | Authors: | Huegle, M, Wohlwend, D. | | Deposit date: | 2017-04-28 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Beyond the BET Family: Targeting CBP/p300 with 4-Acyl Pyrroles.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5NU5
 
 | |
5LPJ
 
 | |
5LPK
 
 | |
5LPL
 
 | | Crystal structure of the bromodomain of human CREBBP bound to the inhibitor XDM3c | | Descriptor: | CREB-binding protein, ~{N}-[(1~{R},2~{R})-7-chloranyl-2-oxidanyl-1,2,3,4-tetrahydronaphthalen-1-yl]-4-ethanoyl-3-ethyl-5-methyl-1~{H}-pyrrole-2-carboxamide | | Authors: | Wohlwend, D, Huegle, M. | | Deposit date: | 2016-08-13 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Beyond the BET Family: Targeting CBP/p300 with 4-Acyl Pyrroles.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5LPM
 
 | | Crystal structure of the bromodomain of human Ep300 bound to the inhibitor XDM3d | | Descriptor: | ACETATE ION, Histone acetyltransferase p300, ~{N}-[(1~{S},2~{S})-7-chloranyl-2-oxidanyl-1,2,3,4-tetrahydronaphthalen-1-yl]-4-ethanoyl-3-ethyl-5-methyl-1~{H}-pyrrole -2-carboxamide | | Authors: | Huegle, M, Wohlwend, D, Gerhardt, S. | | Deposit date: | 2016-08-13 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Beyond the BET Family: Targeting CBP/p300 with 4-Acyl Pyrroles.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6RWJ
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUG0 (6) | | Descriptor: | Bromodomain-containing protein 4, ~{N},3-dimethyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-06-05 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
6S4B
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX1 (8) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Bromodomain-containing protein 4, CALCIUM ION, ... | | Authors: | Huegle, M. | | Deposit date: | 2019-06-27 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
6S6K
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX2 (9) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Bromodomain-containing protein 4, ~{N}-[5-(azepan-1-ylsulfonyl)-2-methoxy-phenyl]-3-methyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-07-03 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
6SA2
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX3 (10) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-methoxy-5-morpholin-4-ylsulfonyl-phenyl)-3-methyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
6SAH
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX5 (11) | | Descriptor: | Bromodomain-containing protein 4, ~{N}-(2-methoxy-5-piperidin-1-ylsulfonyl-phenyl)-3-methyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
6SA3
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX4 (13) | | Descriptor: | Bromodomain-containing protein 4, ~{N}-[2-methoxy-5-(4-methylpiperazin-1-yl)sulfonyl-phenyl]-3-methyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
6SB8
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX14 (7) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Bromodomain-containing protein 4, ~{N}-[5-(diethylsulfamoyl)-2-oxidanyl-phenyl]-3-methyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-07-19 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
6SAJ
 
 | | Crystal Structure of BRD4(1) bound to inhibitor BUX6 (12) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Bromodomain-containing protein 4, ~{N}-[2-methoxy-5-(2-oxa-6-azaspiro[3.3]heptan-6-ylsulfonyl)phenyl]-3-methyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | Authors: | Huegle, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
5D3J
 
 | | First bromodomain of BRD4 bound to inhibitor XD33 | | Descriptor: | 4-acetyl-N-[3-(diethylsulfamoyl)phenyl]-3-ethyl-5-methyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Wohlwend, D, Huegle, M. | | Deposit date: | 2015-08-06 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
