4URF
 
 | | Molecular Genetic and Crystal Structural Analysis of 1-(4- Hydroxyphenyl)-Ethanol Dehydrogenase from Aromatoleum aromaticum EbN1 | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ACETATE ION, BICARBONATE ION, ... | | Authors: | Buesing, I, Hoeffken, H.W, Breuer, M, Woehlbrand, L, Hauer, B, Rabus, R. | | Deposit date: | 2014-06-28 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular Genetic and Crystal Structural Analysis of 1-(4-Hydroxyphenyl)-Ethanol Dehydrogenase from 'Aromatoleum Aromaticum' Ebn1.
J.Mol.Microbiol.Biotechnol., 25, 2015
|
|
4V9S
 
 | | Crystal structure of antibiotic GE82832 bound to 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
4URE
 
 | | Molecular Genetic and Crystal Structural Analysis of 1-(4- Hydroxyphenyl)-Ethanol Dehydrogenase from Aromatoleum aromaticum EbN1 | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ACETATE ION, CYCLOHEXANOL DEHYDROGENASE, ... | | Authors: | Buesing, I, Hoeffken, H.W, Breuer, M, Woehlbrand, L, Hauer, B, Rabus, R. | | Deposit date: | 2014-06-28 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Genetic and Crystal Structural Analysis of 1-(4-Hydroxyphenyl)-Ethanol Dehydrogenase from 'Aromatoleum Aromaticum' Ebn1.
J.Mol.Microbiol.Biotechnol., 25, 2015
|
|
4V9R
 
 | | Crystal structure of antibiotic DITYROMYCIN bound to 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
4DX6
 
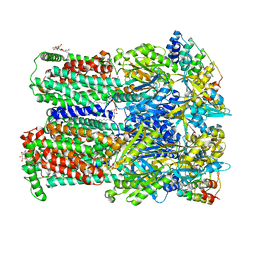 | | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop | | Descriptor: | Acriflavine resistance protein B, DARPIN, DODECYL-BETA-D-MALTOSIDE | | Authors: | Eicher, T, Cha, H, Seeger, M.A, Brandstaetter, L, El-Delik, J, Bohnert, J.A, Kern, W.V, Verrey, F, Gruetter, M.G, Diederichs, K, Pos, K.M. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DX7
 
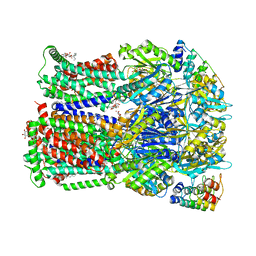 | | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop | | Descriptor: | Acriflavine resistance protein B, DARPIN, DECANE, ... | | Authors: | Eicher, T, Cha, H, Seeger, M.A, Brandstaetter, L, El-Delik, J, Bohnert, J.A, Kern, W.V, Verrey, F, Gruetter, M.G, Diederichs, K, Pos, K.M. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DX5
 
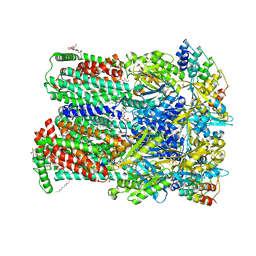 | | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, Acriflavine resistance protein B, DARPIN, ... | | Authors: | Eicher, T, Cha, H, Seeger, M.A, Brandstaetter, L, El-Delik, J, Bohnert, J.A, Kern, W.V, Verrey, F, Gruetter, M.G, Diederichs, K, Pos, K.M. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5ME0
 
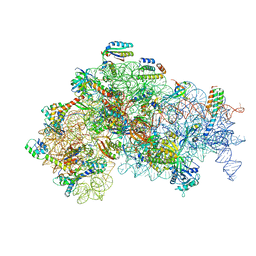 | | Structure of the 30S Pre-Initiation Complex 1 (30S IC-1) Stalled by GE81112 | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Fabbretti, A, Kaminishi, T, Iturrioz, I, Brandi, L, Gil Carton, D, Gualerzi, C, Fucini, P, Connell, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-01-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Structure of a 30S pre-initiation complex stalled by GE81112 reveals structural parallels in bacterial and eukaryotic protein synthesis initiation pathways.
Nucleic Acids Res., 45, 2017
|
|
5ME1
 
 | | Structure of the 30S Pre-Initiation Complex 2 (30S IC-2) Stalled by GE81112 | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Fabbretti, A, Kaminishi, T, Iturrioz, I, Brandi, L, Gil Carton, D, Gualerzi, C, Fucini, P, Connell, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-01-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Structure of a 30S pre-initiation complex stalled by GE81112 reveals structural parallels in bacterial and eukaryotic protein synthesis initiation pathways.
Nucleic Acids Res., 45, 2017
|
|
3ZQ3
 
 | | Crystal Structure of Rat Odorant Binding Protein 3 (OBP3) | | Descriptor: | OBP3 PROTEIN | | Authors: | Portman, K.L, Long, J, Carr, S, Brand, L, Winzor, D.J, Searle, M, Scott, D.J. | | Deposit date: | 2013-03-05 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enthalpy/Entropy Compensation Effects from Cavity Desolvation Underpin Broad Ligand Binding Selectivity for Rat Odorant Binding Protein 3
Biochemistry, 53, 2014
|
|
4U95
 
 | | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, DARPin, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pos, K.M. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB
eLife, 3, 2014
|
|
4U8V
 
 | | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, DARPin, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pos, K.M. | | Deposit date: | 2014-08-04 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB.
Elife, 3, 2014
|
|
4U96
 
 | |
4U8Y
 
 | | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, DARPin, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pos, K.M. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB.
Elife, 3, 2014
|
|
5IWA
 
 | | Crystal structure of the 30S ribosomal subunit from Thermus thermophilus in complex with the GE81112 peptide antibiotic | | Descriptor: | (2S,3S)-2-{[(2S)-3-(2-amino-1H-imidazol-5-yl)-2-{[(2S,4S)-5-(carbamoyloxy)-4-hydroxy-2-({[(2S,3S)-3-hydroxypiperidin-2-yl]carbonyl}amino)pentanoyl]amino}propanoyl]amino}-3-(2-chloro-1H-imidazol-5-yl)-3-hydroxypropanoic acid, 16S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Schedlbauer, A, Kaminishi, T, Ochoa-Lizarralde, B, Chieko, N, Masahito, K, Takemoto, C, Yokoyama, S, Connell, S.R, Fucini, P. | | Deposit date: | 2016-03-22 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of translation initiation complex formation by GE81112 unravels a 16S rRNA structural switch involved in P-site decoding.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5DM6
 
 | | Crystal structure of the 50S ribosomal subunit from Deinococcus radiodurans | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | Authors: | Kaminishi, T, Schedlbauer, A, Ochoa-Lizarralde, B, Connell, S.R, Fucini, P. | | Deposit date: | 2015-09-08 | | Release date: | 2015-11-11 | | Last modified: | 2016-02-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic characterization of the ribosomal binding site and molecular mechanism of action of Hygromycin A.
Nucleic Acids Res., 43, 2015
|
|
5DM7
 
 | | Crystal structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with hygromycin A | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | Authors: | Kaminishi, T, Schedlbauer, A, Ochoa-Lizarralde, B, Connell, S.R, Fucini, P. | | Deposit date: | 2015-09-08 | | Release date: | 2015-11-11 | | Last modified: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic characterization of the ribosomal binding site and molecular mechanism of action of Hygromycin A.
Nucleic Acids Res., 43, 2015
|
|
