6I3N
 
 | |
7OI3
 
 | | Cryo-EM structure of the Cetacean morbillivirus nucleoprotein-RNA complex | | Descriptor: | Cetacean morbillivirus nucleoprotein, poly-A 6-mer | | Authors: | Zinzula, L, Beck, F, Klumpe, S, Bohn, S, Pfeifer, G, Bollschweiler, D, Nagy, I, Plitzko, J.M, Baumeister, W. | | Deposit date: | 2021-05-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the cetacean morbillivirus nucleoprotein-RNA complex.
J.Struct.Biol., 213, 2021
|
|
8PKI
 
 | | Cryo-EM structure of NR5A2-nucleosome complex SHL+5.5 | | Descriptor: | DNA, Histone H2A, Histone H2B type 1-C/E/G, ... | | Authors: | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8PKJ
 
 | | Cryo-EM structure of the nucleosome containing Nr5a2 motif at SHL+5.5 | | Descriptor: | DNA, Histone H2A, Histone H2B, ... | | Authors: | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8BQ2
 
 | | CryoEM structure of the pre-synaptic RAD51 nucleoprotein filament in the presence of ATP and Ca2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (30-MER), ... | | Authors: | Appleby, R, Bollschweiler, D, Pellegrini, L. | | Deposit date: | 2022-11-18 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A metal ion-dependent mechanism of RAD51 nucleoprotein filament disassembly.
Iscience, 26, 2023
|
|
8BSC
 
 | |
8BR2
 
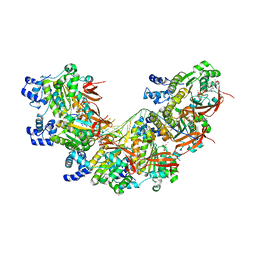 | | CryoEM structure of the post-synaptic RAD51 nucleoprotein filament in the presence of ATP and Ca2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*GP*CP*GP*AP*GP*CP*TP*CP*GP*AP*TP*GP*CP*AP*CP*CP*TP*CP*CP*A)-3'), ... | | Authors: | Appleby, R, Bollschweiler, D, Pellegrini, L. | | Deposit date: | 2022-11-22 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A metal ion-dependent mechanism of RAD51 nucleoprotein filament disassembly.
Iscience, 26, 2023
|
|
6Z6F
 
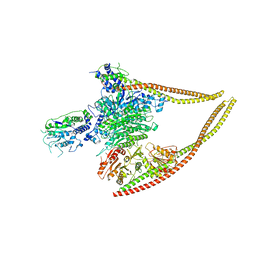 | | HDAC-PC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6H
 
 | | HDAC-DC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (8.55 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6O
 
 | | HDAC-TC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6P
 
 | | HDAC-PC-Nuc | | Descriptor: | DNA (145-MER), HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6SWY
 
 | | Structure of active GID E3 ubiquitin ligase complex minus Gid2 and delta Gid9 RING domain | | Descriptor: | Glucose-induced degradation protein 8, Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10, Vacuolar import and degradation protein 24, ... | | Authors: | Qiao, S, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2019-09-24 | | Release date: | 2019-11-20 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Interconversion between Anticipatory and Active GID E3 Ubiquitin Ligase Conformations via Metabolically Driven Substrate Receptor Assembly
Mol.Cell, 77, 2020
|
|
5EQW
 
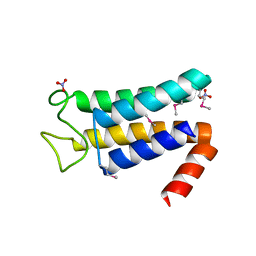 | | Structure of the major structural protein D135 of Acidianus tailed spindle virus (ATSV) | | Descriptor: | NITRATE ION, Putative major coat protein | | Authors: | Hochstein, R.A, Lintner, N.G, Young, M.J, Lawrence, C.M. | | Deposit date: | 2015-11-13 | | Release date: | 2016-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Structural studies ofAcidianustailed spindle virus reveal a structural paradigm used in the assembly of spindle-shaped viruses.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6YBQ
 
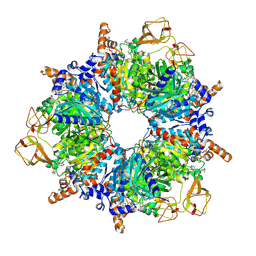 | | Engineered glycolyl-CoA carboxylase (quintuple mutant) with bound CoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Propionyl-CoA carboxylase alpha subunit, ... | | Authors: | Schuller, J.M, Schuller, S.K, Zarzycki, J, Scheffen, M, Marchal, D.M, Erb, T.J. | | Deposit date: | 2020-03-17 | | Release date: | 2020-10-28 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (1.96 Å) | | Cite: | A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation
Nat Catal, 2021
|
|
6YBP
 
 | | Propionyl-CoA carboxylase of Methylorubrum extorquens with bound CoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Propionyl-CoA carboxylase alpha subunit, ... | | Authors: | Schuller, J.M, Schuller, S.K, Zarzycki, J, Scheffen, M, Marchal, D.M, Erb, T.J. | | Deposit date: | 2020-03-17 | | Release date: | 2020-10-28 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation
Nat Catal, 2021
|
|
