4DIN
 
 | | Novel Localization and Quaternary Structure of the PKA RI beta Holoenzyme | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Ilouz, R, Bubis, J, Wu, J, Yim, Y.Y, Deal, M.S, Kornev, A.P, Ma, Y, Blumenthal, D.K, Taylor, S.S. | | Deposit date: | 2012-01-31 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Localization and quaternary structure of the PKA RI Beta holoenzyme
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4MX3
 
 | | Crystal Structure of PKA RIalpha Homodimer | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Bruystens, J.G.H, Wu, J, Fortezzo, A, Kornev, A.P, Blumenthal, D.A, Taylor, S.S. | | Deposit date: | 2013-09-25 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.88 Å) | | Cite: | PKA RI alpha Homodimer Structure Reveals an Intermolecular Interface with Implications for Cooperative cAMP Binding and Carney Complex Disease.
Structure, 22, 2014
|
|
5JR7
 
 | | Crystal structure of an ACRDYS heterodimer [RIa(92-365):C] of PKA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Bruystens, J.G.H, Wu, J, Taylor, S.S. | | Deposit date: | 2016-05-05 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Structure of a PKA RI alpha Recurrent Acrodysostosis Mutant Explains Defective cAMP-Dependent Activation.
J. Mol. Biol., 428, 2016
|
|
6O5V
 
 | | Binary complex of native hAChE with oxime reactivator RS-170B | | Descriptor: | 4-carbamoyl-1-(3-{2-[(E)-(hydroxyimino)methyl]-1H-imidazol-1-yl}propyl)pyridin-1-ium, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Gerlits, O, Kovalevsky, A, Radic, Z. | | Deposit date: | 2019-03-04 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Productive reorientation of a bound oxime reactivator revealed in room temperature X-ray structures of native and VX-inhibited human acetylcholinesterase.
J.Biol.Chem., 294, 2019
|
|
6O66
 
 | |
6U37
 
 | | Structure of VX-phosphonylated hAChE in complex with oxime reactivator RS194B | | Descriptor: | (2E)-N-[2-(azepan-1-yl)ethyl]-2-(hydroxyimino)acetamide, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Rational design, synthesis, and evaluation of uncharged, "smart" bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase.
J.Biol.Chem., 295, 2020
|
|
6O5R
 
 | |
6U3P
 
 | | Binary complex of native hAChE with oxime reactivator LG-703 | | Descriptor: | (2E,2'E)-N,N'-[1,4-diazepane-1,4-diyldi(ethane-2,1-diyl)]bis[2-(hydroxyimino)acetamide], Acetylcholinesterase, DIMETHYL SULFOXIDE | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2019-08-22 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Rational design, synthesis, and evaluation of uncharged, "smart" bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase.
J.Biol.Chem., 295, 2020
|
|
6O5S
 
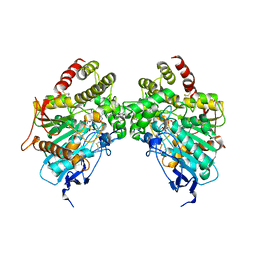 | |
6U34
 
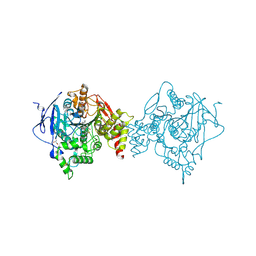 | | Binary complex of native hAChE with oxime reactivator RS194B | | Descriptor: | (2E)-N-[2-(azepan-1-yl)ethyl]-2-(hydroxyimino)acetamide, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rational design, synthesis, and evaluation of uncharged, "smart" bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase.
J.Biol.Chem., 295, 2020
|
|
8DT4
 
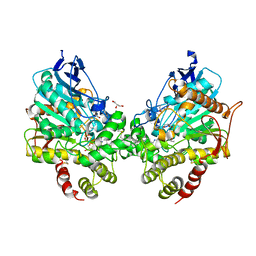 | | X-ray structure of human acetylcholinesterase ternary complex with paraoxon and oxime MMB4 (POX-hAChE-MMB4) | | Descriptor: | 1,1'-methylenebis{4-[(E)-(hydroxyimino)methyl]pyridin-1-ium}, Acetylcholinesterase, DIETHYL PHOSPHONATE, ... | | Authors: | Kovalevsky, A.Y, Gerlits, O, Radic, Z. | | Deposit date: | 2022-07-25 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and dynamic effects of paraoxon binding to human acetylcholinesterase by X-ray crystallography and inelastic neutron scattering.
Structure, 30, 2022
|
|
8DT2
 
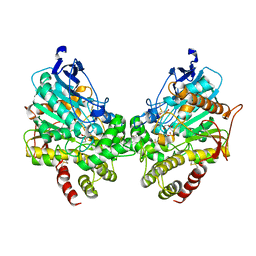 | | X-ray structure of human acetylcholinesterase inhibited by paraoxon (POX-hAChE) | | Descriptor: | Acetylcholinesterase, DIETHYL PHOSPHONATE, DIMETHYL SULFOXIDE, ... | | Authors: | Kovalevsky, A.Y, Gerlits, O, Radic, Z. | | Deposit date: | 2022-07-25 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structural and dynamic effects of paraoxon binding to human acetylcholinesterase by X-ray crystallography and inelastic neutron scattering.
Structure, 30, 2022
|
|
8DT7
 
 | | X-ray structure of human acetylcholinesterase in complex with oxime MMB4 (hAChE-MMB4) | | Descriptor: | 1,1'-methylenebis{4-[(E)-(hydroxyimino)methyl]pyridin-1-ium}, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Kovalevsky, A.Y, Gerlits, O, Radic, Z. | | Deposit date: | 2022-07-25 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.207 Å) | | Cite: | Structural and dynamic effects of paraoxon binding to human acetylcholinesterase by X-ray crystallography and inelastic neutron scattering.
Structure, 30, 2022
|
|
8DT5
 
 | | X-ray structure of human acetylcholinesterase ternary complex with paraoxon and oxime RS170B (POX-hAChE-RS170B) | | Descriptor: | 4-carbamoyl-1-(3-{2-[(E)-(hydroxyimino)methyl]-1H-imidazol-1-yl}propyl)pyridin-1-ium, Acetylcholinesterase, DIETHYL PHOSPHONATE, ... | | Authors: | Kovalevsky, A.Y, Gerlits, O, Radic, Z. | | Deposit date: | 2022-07-25 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and dynamic effects of paraoxon binding to human acetylcholinesterase by X-ray crystallography and inelastic neutron scattering.
Structure, 30, 2022
|
|
1DEG
 
 | | THE LINKER OF DES-GLU84 CALMODULIN IS BENT AS SEEN IN THE CRYSTAL STRUCTURE | | Descriptor: | CALCIUM ION, CALMODULIN | | Authors: | Raghunathan, S, Chandross, R, Cheng, B.P, Persechini, A, Sobottk, S.E, Kretsinger, R.H. | | Deposit date: | 1993-06-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The linker of des-Glu84-calmodulin is bent.
Proc.Natl.Acad.Sci.Usa, 90, 1993
|
|
