5OAU
 
 | |
5IRP
 
 | | Crystal structure of the alanine racemase Bsu17640 from Bacillus subtilis | | Descriptor: | (5-hydroxy-6-methylpyridin-3-yl)methyl dihydrogen phosphate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alanine racemase 2, ... | | Authors: | Bernardo-Garcia, N, Gago, F, Hermoso, J.A. | | Deposit date: | 2016-03-14 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cold-induced aldimine bond cleavage by Tris in Bacillus subtilis alanine racemase.
Org.Biomol.Chem., 17, 2019
|
|
5EHD
 
 | | Crystal structure of human nucleophosmin-core in complex with cytochrome c | | Descriptor: | CHLORIDE ION, Nucleophosmin | | Authors: | Bernardo-Garcia, N, Hermoso, J.A, Gonzalez-Arzola, K, Diaz-Moreno, I, De la Rosa, M.A. | | Deposit date: | 2015-10-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of human nucleophosmin-core in complex with cytochrome c
To Be Published
|
|
6Q70
 
 | | Crystal structure of the alanine racemase Bsu17640 from Bacillus subtilis in the presence of HEPES | | Descriptor: | Alanine racemase 2, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bernardo-Garcia, N, Gago, F, Hermoso, J.A. | | Deposit date: | 2018-12-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cold-induced aldimine bond cleavage by Tris in Bacillus subtilis alanine racemase.
Org.Biomol.Chem., 17, 2019
|
|
6Q71
 
 | | Crystal structure of the alanine racemase Bsu17640 from Bacillus subtilis in the presence of Bis-Tris propane | | Descriptor: | Alanine racemase 2, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bernardo-Garcia, N, Gago, F, Hermoso, J.A. | | Deposit date: | 2018-12-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Cold-induced aldimine bond cleavage by Tris in Bacillus subtilis alanine racemase.
Org.Biomol.Chem., 17, 2019
|
|
6Q72
 
 | |
5OJ1
 
 | | Penicillin Binding Protein 2x (PBP2x) from S.pneumoniae in complex with Oxacillin and a tetrasaccharide | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Penicillin-binding protein 2X, SODIUM ION | | Authors: | Bernardo-Garcia, N, Hermoso, J.A. | | Deposit date: | 2017-07-20 | | Release date: | 2018-05-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Allostery, Recognition of Nascent Peptidoglycan, and Cross-linking of the Cell Wall by the Essential Penicillin-Binding Protein 2x of Streptococcus pneumoniae.
ACS Chem. Biol., 13, 2018
|
|
5OIZ
 
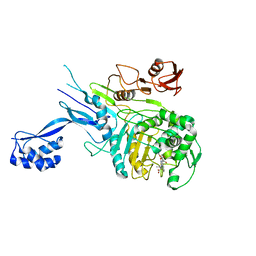 | | Penicillin-Binding Protein 2X (PBP2X) from Streptococcus pneumoniae in complex with oxacillin | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Penicillin-binding protein 2X | | Authors: | Bernardo-Garcia, N, Hermoso, J.A. | | Deposit date: | 2017-07-20 | | Release date: | 2018-05-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allostery, Recognition of Nascent Peptidoglycan, and Cross-linking of the Cell Wall by the Essential Penicillin-Binding Protein 2x of Streptococcus pneumoniae.
ACS Chem. Biol., 13, 2018
|
|
5OJ0
 
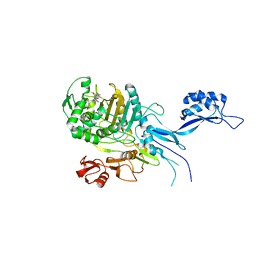 | |
5I8L
 
 | | Crystal structure of the full-length cell wall-binding module of Cpl7 mutant R223A | | Descriptor: | GLYCEROL, Lysozyme | | Authors: | Bernardo-Garcia, N, Silva-Martin, N, Uson, I, Hermoso, J.A. | | Deposit date: | 2016-02-19 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Deciphering how Cpl-7 cell wall-binding repeats recognize the bacterial peptidoglycan.
Sci Rep, 7, 2017
|
|
7QBS
 
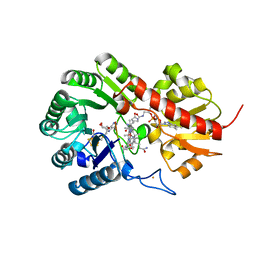 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, S-adenosyl-L-methionine, and peptide bound. | | Descriptor: | CO-METHYLCOBALAMIN, FE (III) ION, IRON/SULFUR CLUSTER, ... | | Authors: | Bernardo-Garcia, N, Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
3ZLB
 
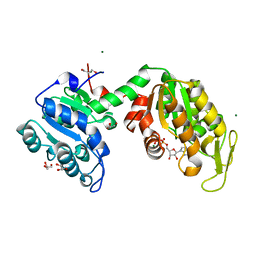 | |
4CFP
 
 | | Crystal structure of MltC in complex with tetrasaccharide at 2.15 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, CITRIC ACID, HEXAETHYLENE GLYCOL, ... | | Authors: | Bernardo-Garcia, N, Artola-Recolons, C, Hermoso, J.A. | | Deposit date: | 2013-11-19 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Cell Wall Cleavage by Modular Lytic Transglycosylase Mltc of Escherichia Coli.
Acs Chem.Biol., 9, 2014
|
|
4BHY
 
 | | Structure of alanine racemase from Aeromonas hydrophila | | Descriptor: | ALANINE RACEMASE | | Authors: | Otero, L.H, Carrasco-Lopez, C, Bernardo-Garcia, N, Hermoso, J.A. | | Deposit date: | 2013-04-09 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural Basis for the Broad Specificity of a New Family of Amino-Acid Racemases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CFO
 
 | | Structure of Lytic Transglycosylase MltC from Escherichia coli in complex with tetrasaccharide at 2.9 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-3-O-[(2R)-1-amino-1-oxopropan-2-yl]-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(2R)-1-amino-1-oxopropan-2-yl]-2-deoxy-beta-D-glucopyranoside, MLTC | | Authors: | Artola-Recolons, C, Bernardo-Garcia, N, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2013-11-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and Cell Wall Cleavage by Modular Lytic Transglycosylase Mltc of Escherichia Coli.
Acs Chem.Biol., 9, 2014
|
|
4C5F
 
 | |
4CHX
 
 | | Crystal structure of MltC in complex with disaccharide pentapeptide DHl89 | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Membrane-bound lytic murein transglycosylase C, ... | | Authors: | Artola-Recolons, C, Bernardo-Garcia, N, Hermoso, J.A. | | Deposit date: | 2013-12-04 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure and Cell Wall Cleavage by Modular Lytic Transglycosylase Mltc of Escherichia Coli.
Acs Chem.Biol., 9, 2014
|
|
4CVD
 
 | |
7AGZ
 
 | | BsrV no-histagged | | Descriptor: | Broad specificity amino-acid racemase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Espaillat, A, Cava, F, Hermoso, J.A. | | Deposit date: | 2020-09-23 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Binding of non-canonical peptidoglycan controls Vibrio cholerae broad spectrum racemase activity.
Comput Struct Biotechnol J, 19, 2021
|
|
7QBU
 
 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, and S-methyl-5'-thioadenosine bound. | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CO-METHYLCOBALAMIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
7QBT
 
 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, and S-methyl-5'-thioadenosine bound. | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CO-METHYLCOBALAMIN, FE (III) ION, ... | | Authors: | Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
7QBV
 
 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, and S-adenosyl-L-homocysteine bound. | | Descriptor: | CO-METHYLCOBALAMIN, FE (III) ION, IRON/SULFUR CLUSTER, ... | | Authors: | Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
4BF5
 
 | |
4BEU
 
 | |
4BEQ
 
 | |
