7K5F
 
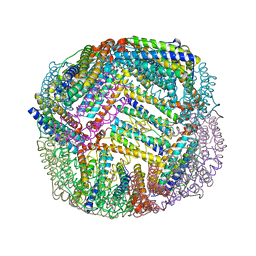 | | 1.90 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor KM-5-50 | | 分子名称: | 4-{[(3-chloro-5-hydroxyphenyl)methyl]amino}-1H-isoindole-1,3(2H)-dione, Ferroxidase, POTASSIUM ION, ... | | 著者 | Lovell, S, Battaile, K.P, Soldano, A, Punchi-Hewage, A, Meraz, K, Annor-Gyamfi, J.K, Yao, H, Bunce, R.A, Rivera, M. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Small Molecule Inhibitors of the Bacterioferritin (BfrB)-Ferredoxin (Bfd) Complex Kill Biofilm-Embedded Pseudomonas aeruginosa Cells.
Acs Infect Dis., 7, 2021
|
|
7K7B
 
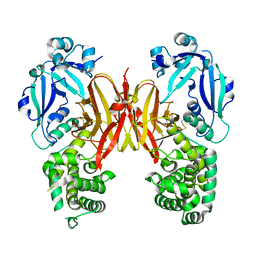 | | Crystal structure of diphtheria toxin from crystals obtained at pH 5.0 | | 分子名称: | Diphtheria toxin | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rodnin, M.V, Ladokhin, A.S. | | 登録日 | 2020-09-22 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure of the Diphtheria Toxin at Acidic pH: Implications for the Conformational Switching of the Translocation Domain.
Toxins, 12, 2020
|
|
7K5G
 
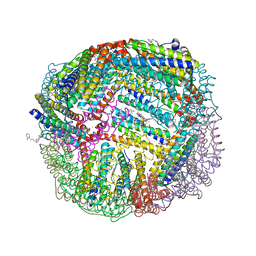 | | 1.95 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor KM-5-28 | | 分子名称: | 4-{[3-(2-hydroxyphenyl)propyl]amino}-1H-isoindole-1,3(2H)-dione, Ferroxidase, POTASSIUM ION, ... | | 著者 | Lovell, S, Battaile, K.P, Soldano, A, Punchi-Hewage, A, Meraz, K, Annor-Gyamfi, J.K, Yao, H, Bunce, R.A, Rivera, M. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Small Molecule Inhibitors of the Bacterioferritin (BfrB)-Ferredoxin (Bfd) Complex Kill Biofilm-Embedded Pseudomonas aeruginosa Cells.
Acs Infect Dis., 7, 2021
|
|
7K7C
 
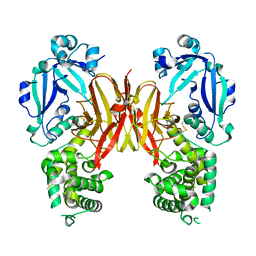 | | Crystal structure of diphtheria toxin from crystals obtained at pH 5.5 | | 分子名称: | Diphtheria toxin | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rodnin, M.V, Ladokhin, A.S. | | 登録日 | 2020-09-22 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure of the Diphtheria Toxin at Acidic pH: Implications for the Conformational Switching of the Translocation Domain.
Toxins, 12, 2020
|
|
7K0G
 
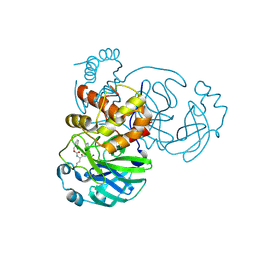 | | 1.85 A resolution structure of SARS-CoV 3CL protease in complex with deuterated GC376 | | 分子名称: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Replicase polyprotein 1a | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Nguyen, H.N, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-09-04 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K0H
 
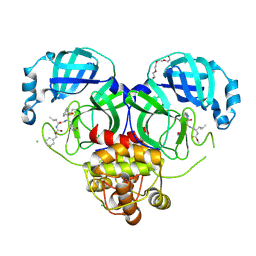 | | 1.70 A resolution structure of SARS-CoV 3CL protease in complex with a deuterated GC376 alpha-ketoamide analog (compound 5) | | 分子名称: | CHLORIDE ION, N-{(2S,3R)-4-(benzylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-N~2~-[(benzyloxy)carbonyl]-L-leucinamide, Replicase polyprotein 1a, ... | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Nguyen, H.N, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-09-04 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K0E
 
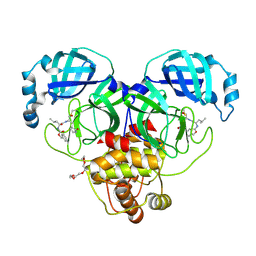 | | 1.90 A resolution structure of SARS-CoV-2 3CL protease in complex with deuterated GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, TETRAETHYLENE GLYCOL | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Nguyen, H.N, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-09-04 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8TGB
 
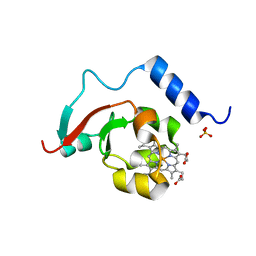 | | Crystal structure of root lateral formation protein (RLF) b5-domain from Oryza sativa | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, root lateral formation protein (RLF) | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Benson, D.R. | | 登録日 | 2023-07-12 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The N-terminal intrinsically disordered region of Ncb5or docks with the cytochrome b 5 core to form a helical motif that is of ancient origin.
Proteins, 92, 2024
|
|
8UA2
 
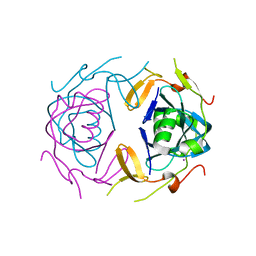 | | Crystal Structure of infected cell protein 0 (ICP0) from herpes simplex virus 1 (proteolyzed fragment) | | 分子名称: | IODIDE ION, RL2 | | 著者 | Lovell, S, Kashipathy, M, Battaile, K.P, Cooper, A, Davido, D. | | 登録日 | 2023-09-20 | | 公開日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | HSV-1 ICP0 dimer domain adopts a novel beta-barrel fold.
Proteins, 2024
|
|
8UA5
 
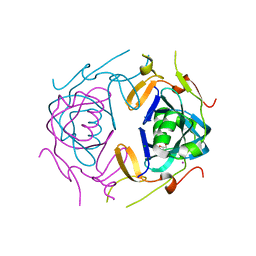 | | Crystal Structure of infected cell protein 0 (ICP0) from herpes simplex virus 1 (A636-Q776) | | 分子名称: | CHLORIDE ION, GLYCEROL, IODIDE ION, ... | | 著者 | Lovell, S, Kashipathy, M, Battaile, K.P, Cooper, A, Davido, D. | | 登録日 | 2023-09-20 | | 公開日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | HSV-1 ICP0 dimer domain adopts a novel beta-barrel fold.
Proteins, 2024
|
|
8G1W
 
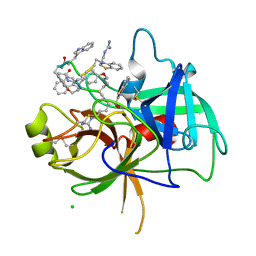 | | Crystal Structure Matriptase (C731S) in Complex with Inhibitor VD4162B | | 分子名称: | CHLORIDE ION, Cyclic peptide inhibitor (ACE)Y(DTR)(NLE)(KCM), Cyclic peptide inhibitor (ACE)Y(DTR)(NLE)(THZ), ... | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Janetka, J.W. | | 登録日 | 2023-02-03 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Mechanism-Based Macrocyclic Inhibitors of Serine Proteases.
J.Med.Chem., 67, 2024
|
|
6P8I
 
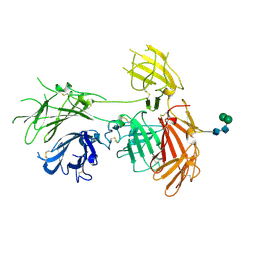 | | N-terminal 5 domains of IGFIIR | | 分子名称: | Cation-independent mannose-6-phosphate receptor, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Olson, L.J, Dahms, N.M, Kim, J.-J.P. | | 登録日 | 2019-06-07 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol, 3, 2020
|
|
2OB9
 
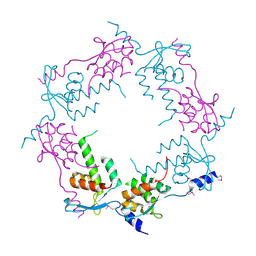 | | Structure of bacteriophage HK97 tail assembly chaperone | | 分子名称: | Tail assembly chaperone | | 著者 | McGrath, T.E, Tuite, A, Bona, D, Saridakis, V, Edwards, A.M, Maxwell, K, Chirgadze, N.Y. | | 登録日 | 2006-12-18 | | 公開日 | 2007-12-18 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A conserved spiral structure for highly diverged phage tail assembly chaperones.
J.Mol.Biol., 425, 2013
|
|
9BN8
 
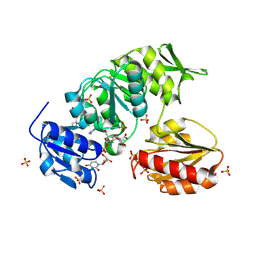 | |
9BN9
 
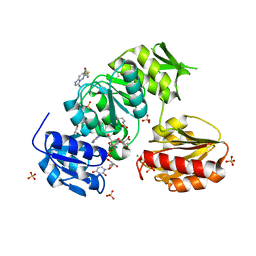 | |
3D4J
 
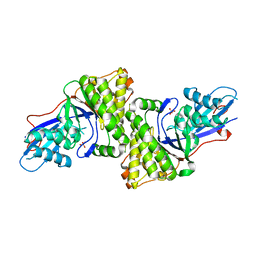 | | Crystal structure of Human mevalonate diphosphate decarboxylase | | 分子名称: | Diphosphomevalonate decarboxylase, SULFATE ION | | 著者 | Voynova, N.E, Fu, Z, Battaile, K, Herdendorf, T.J, Kim, J.-J.P, Miziorko, H.M. | | 登録日 | 2008-05-14 | | 公開日 | 2008-12-16 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Human mevalonate diphosphate decarboxylase: characterization, investigation of the mevalonate diphosphate binding site, and crystal structure.
Arch.Biochem.Biophys., 480, 2008
|
|
1Y0O
 
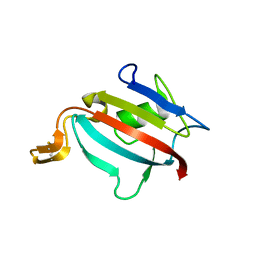 | |
5KGN
 
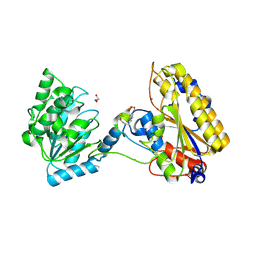 | | 1.95A resolution structure of independent phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (2d) | | 分子名称: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, GLYCEROL, ... | | 著者 | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | 登録日 | 2016-06-13 | | 公開日 | 2017-04-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
5KGM
 
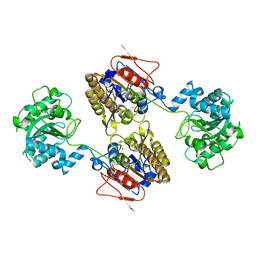 | | 2.95A resolution structure of Apo independent phosphoglycerate mutase from C. elegans (monoclinic form) | | 分子名称: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, MANGANESE (II) ION, ... | | 著者 | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | 登録日 | 2016-06-13 | | 公開日 | 2017-04-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
5KGL
 
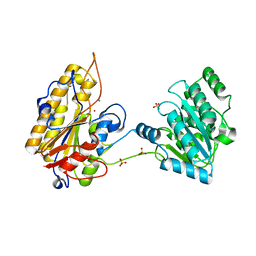 | | 2.45A resolution structure of Apo independent phosphoglycerate mutase from C. elegans (orthorhombic form) | | 分子名称: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, MANGANESE (II) ION, ... | | 著者 | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | 登録日 | 2016-06-13 | | 公開日 | 2017-04-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
2Z5D
 
 | | Human ubiquitin-conjugating enzyme E2 H | | 分子名称: | SODIUM ION, Ubiquitin-conjugating enzyme E2 H | | 著者 | Bochkarev, A, Cui, H, Walker, J.R, Newman, E.M, Mackenzie, F, Battaile, K.P, Sundstrom, M, Arrowsmith, C, Edwards, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | 登録日 | 2007-07-06 | | 公開日 | 2007-10-09 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
MOL.CELL PROTEOMICS, 11, 2012
|
|
3CXN
 
 | | Structure of the Urease Accessory Protein UreF from Helicobacter pylori | | 分子名称: | GLYCEROL, Urease accessory protein ureF | | 著者 | Lam, R, Johns, K, Romanov, V, Dong, A, Wu-Brown, J, Guthrie, J, Dharamsi, A, Thambipillai, D, Mansoury, K, Edwards, A.M, Pai, E.F, Chirgadze, N.Y. | | 登録日 | 2008-04-24 | | 公開日 | 2009-05-12 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structure of a truncated urease accessory protein UreF from Helicobacter pylori.
Proteins, 78, 2010
|
|
6DE6
 
 | |
6NTY
 
 | | 2.1 A resolution structure of the Musashi-2 (Msi2) RNA recognition motif 1 (RRM1) domain | | 分子名称: | PHOSPHATE ION, RNA-binding protein Musashi homolog 2 | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Lan, L, Xiaoqing, W, Cooper, A, Gao, F.P, Xu, L. | | 登録日 | 2019-01-30 | | 公開日 | 2019-10-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.
Proteins, 88, 2020
|
|
8W21
 
 | |
