5TO3
 
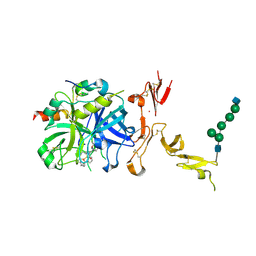 | | Crystal structure of thrombin mutant W215A/E217A fused to EGF456 of thrombomodulin via a 31-residue linker and bound to PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose-(1-4)-[beta-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Barranco-Medina, S, Murphy, M, Pelc, L, Chen, Z, Di Cera, E, Pozzi, N. | | Deposit date: | 2016-10-16 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Rational Design of Protein C Activators.
Sci Rep, 7, 2017
|
|
4RN6
 
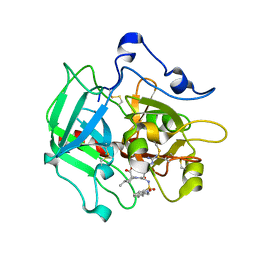 | | Structure of prethrombin-2 mutant s195a bound to the active site inhibitor argatroban | | Descriptor: | (2R,4R)-4-methyl-1-(N~2~-{[(3S)-3-methyl-1,2,3,4-tetrahydroquinolin-8-yl]sulfonyl}-L-arginyl)piperidine-2-carboxylic acid, Thrombin heavy chain | | Authors: | Pozzi, N, Chen, Z, Zapata, F, Niu, W, Barranco-Medina, S, Pelc, L.A, Di Cera, E. | | Deposit date: | 2014-10-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Autoactivation of thrombin precursors.
J.Biol.Chem., 288, 2013
|
|
2PWJ
 
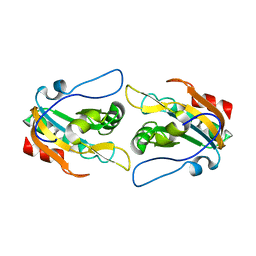 | |
4DT7
 
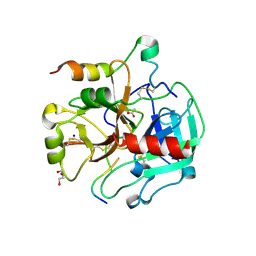 | | Crystal structure of thrombin bound to the activation domain QEDQVDPRLIDGKMTRRGDS of protein C | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Pozzi, N, Barranco-Medina, S, Chen, Z, Di Cera, E. | | Deposit date: | 2012-02-20 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exposure of R169 controls protein C activation and autoactivation.
Blood, 120, 2012
|
|
4HFP
 
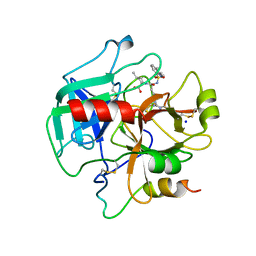 | | Structure of thrombin mutant S195a bound to the active site inhibitor argatroban | | Descriptor: | (2R,4R)-4-methyl-1-(N~2~-{[(3S)-3-methyl-1,2,3,4-tetrahydroquinolin-8-yl]sulfonyl}-L-arginyl)piperidine-2-carboxylic acid, Prothrombin, SODIUM ION | | Authors: | Pozzi, N, Chen, Z, Zapata, F, Lin, W, Barranco-Medina, S, Pelc, L.A, Di Cera, E. | | Deposit date: | 2012-10-05 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Autoactivation of thrombin precursors.
J.Biol.Chem., 288, 2013
|
|
4H6T
 
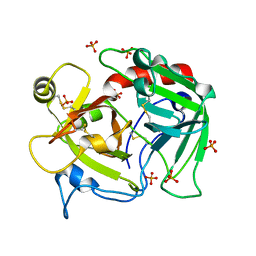 | | Crystal structure of prethrombin-2 mutant E14eA/D14lA/E18A/S195A | | Descriptor: | PHOSPHATE ION, Prothrombin | | Authors: | Pozzi, N, Chen, Z, Zapata, F, Pelc, L.A, Di Cera, E. | | Deposit date: | 2012-09-19 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Autoactivation of thrombin precursors.
J.Biol.Chem., 288, 2013
|
|
4H6S
 
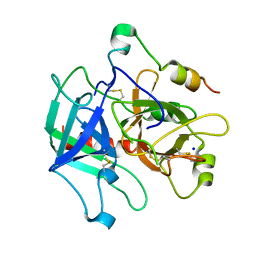 | | Crystal structure of thrombin mutant E14eA/D14lA/E18A/S195A | | Descriptor: | Prothrombin, SODIUM ION | | Authors: | Pozzi, N, Chen, Z, Zapata, F, Pelc, L.A, Di Cera, E. | | Deposit date: | 2012-09-19 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Autoactivation of thrombin precursors.
J.Biol.Chem., 288, 2013
|
|
3SQH
 
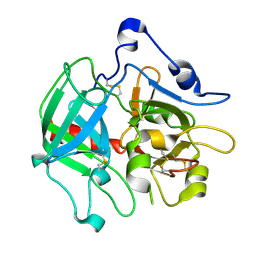 | |
3SQE
 
 | |
