3X21
 
 | | Crystal structure of Escherichia coli nitroreductase NfsB mutant T41L/N71S/F124W | | Descriptor: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Bai, J, Yang, J, Zhou, Y, Yang, Q. | | Deposit date: | 2014-12-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Altering the regioselectivity of a nitroreductase in the synthesis of arylhydroxylamines by structure-based engineering.
Chembiochem, 16, 2015
|
|
3X22
 
 | | Crystal structure of Escherichia coli nitroreductase NfsB mutant N71S/F123A/F124W | | Descriptor: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Bai, J, Yang, J, Zhou, Y, Yang, Q. | | Deposit date: | 2014-12-06 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis of Escherichia coli nitroreductase NfsB triple mutants engineered for improved activity and regioselectivity toward the prodrug CB1954
PROCESS BIOCHEM, 50, 2015
|
|
2Y51
 
 | | Crystal structure of E167A mutant of the box pathway encoded ALDH from Burkholderia xenovorans LB400 | | Descriptor: | ALDEHYDE DEHYDROGENASE (BOX PATHWAY), GLYCEROL | | Authors: | Bains, J, Leon, R, Temke, K.G, Boulanger, M.J. | | Deposit date: | 2011-01-11 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elucidating the Reaction Mechanism of the Benzoate Oxidation Pathway Encoded Aldehyde Dehydrogenase from Burkholderia Xenovorans Lb400.
Protein Sci., 20, 2011
|
|
2XUA
 
 | | Crystal structure of the enol-lactonase from Burkholderia xenovorans LB400 | | Descriptor: | 3-OXOADIPATE ENOL-LACTONASE, LAEVULINIC ACID | | Authors: | Bains, J, Kaufman, L, Farnell, B, Boulanger, M.J. | | Deposit date: | 2010-10-17 | | Release date: | 2011-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Product Analog Bound Form of 3-Oxoadipate-Enol- Lactonase (Pcad) Reveals a Multifunctional Role for the Divergent CAP Domain.
J.Mol.Biol., 406, 2011
|
|
2Y53
 
 | | Crystal structure of E257Q mutant of the box pathway encoded ALDH from Burkholderia xenovorans LB400 | | Descriptor: | ALDEHYDE DEHYDROGENASE (BOX PATHWAY), GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bains, J, Leon, R, Temke, K.G, Boulanger, M.J. | | Deposit date: | 2011-01-11 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Elucidating the Reaction Mechanism of the Benzoate Oxidation Pathway Encoded Aldehyde Dehydrogenase from Burkholderia Xenovorans Lb400.
Protein Sci., 20, 2011
|
|
2Y5D
 
 | | Crystal structure of C296A mutant of the box pathway encoded ALDH from Burkholderia xenovorans LB400 | | Descriptor: | ALDEHYDE DEHYDROGENASE (BOX PATHWAY), GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Bains, J, Leon, R, Temke, K.G, Boulanger, M.J. | | Deposit date: | 2011-01-12 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Elucidating the Reaction Mechanism of the Benzoate Oxidation Pathway Encoded Aldehyde Dehydrogenase from Burkholderia Xenovorans Lb400.
Protein Sci., 20, 2011
|
|
2Y52
 
 | | Crystal structure of E496A mutant of the box pathway encoded ALDH from Burkholderia xenovorans LB400 | | Descriptor: | ALDEHYDE DEHYDROGENASE (BOX PATHWAY), GLYCEROL | | Authors: | Bains, J, Leon, R, Temke, K.G, Boulanger, M.J. | | Deposit date: | 2011-01-11 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Elucidating the Reaction Mechanism of the Benzoate Oxidation Pathway Encoded Aldehyde Dehydrogenase from Burkholderia Xenovorans Lb400.
Protein Sci., 20, 2011
|
|
4ATY
 
 | |
2W3P
 
 | | BoxC crystal structure | | Descriptor: | BENZOYL-COA-DIHYDRODIOL LYASE, BETA-MERCAPTOETHANOL, GLYCEROL | | Authors: | Bains, J, Boulanger, M.J. | | Deposit date: | 2008-11-13 | | Release date: | 2009-04-14 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Biophysical Characterization of Boxc from Burkholderia Xenovorans Lb400: A Novel Ring-Cleaving Enzyme in the Crotonase Superfamily.
J.Biol.Chem., 284, 2009
|
|
2VRO
 
 | | Crystal structure of aldehyde dehydrogenase from Burkholderia xenovorans LB400 | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ALDEHYDE DEHYDROGENASE, HEXAETHYLENE GLYCOL, ... | | Authors: | Bains, J, Boulanger, M.J. | | Deposit date: | 2008-04-09 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Characterization of a Novel Aldehyde Dehydrogenase Encoded by the Benzoate Oxidation (Box) Pathway in Burkholderia Xenovorans Lb400
J.Mol.Biol., 379, 2008
|
|
6FWB
 
 | | Crystal structure of Mat2A at 1.79 Angstron resolution | | Descriptor: | GLYCEROL, S-adenosylmethionine synthase isoform type-2, SODIUM ION, ... | | Authors: | Zhou, A, Wei, Z, Bai, J, Wang, H. | | Deposit date: | 2018-03-06 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of a natural inhibitor of methionine adenosyltransferase 2A regulating one-carbon metabolism in keratinocytes.
Ebiomedicine, 39, 2019
|
|
2V7B
 
 | |
1UU8
 
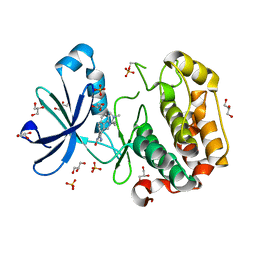 | | Structure of human PDK1 kinase domain in complex with BIM-1 | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, 3-{1-[3-(DIMETHYLAMINO)PROPYL]-1H-INDOL-3-YL}-4-(1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, GLYCEROL, ... | | Authors: | Komander, D, Kular, G.S, Schuttelkopf, A.W, Deak, M, Prakash, K.R, Bain, J, Elliot, M, Garrido-Franco, M, Kozikowski, A.P, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-12-16 | | Release date: | 2004-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interactions of Ly333531 and Other Bisindolyl Maleimide Inhibitors with Pdk1
Structure, 12, 2004
|
|
7M7C
 
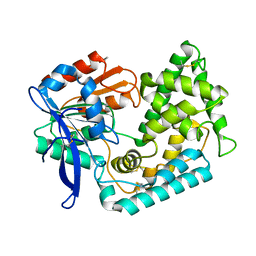 | | Crystal Structure of Hip1 (Rv2224c) mutant - T466A/S228DHA (dehydroalanine) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, Goldfarb, N.E, Doran, M.H, Baikovitz, J, Liu, D, Sun, S, White, A, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitors and Inactivators of Mycobacterium tuberculosis serine protease Hip1 (Rv2224c)
To Be Published
|
|
5BKM
 
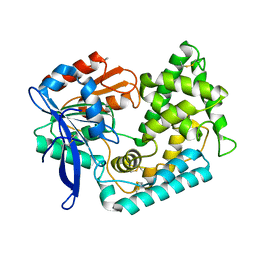 | | Crystal Structure of Hip1 (Rv2224c) mutant - S228DHA (dehydroalanine) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, Goldfarb, N.E, Doran, M.H, Baikovitz, J, Liu, D, Sun, S, White, A, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2021-03-20 | | Release date: | 2022-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal Structure of Hip1 (Rv2224c) mutant - S228DHA (dehydroalanine)
To Be Published
|
|
4WV7
 
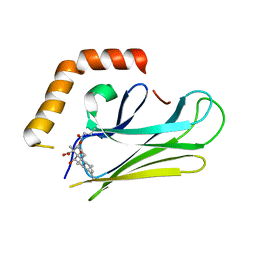 | | HEAT SHOCK PROTEIN 70 SUBSTRATE BINDING DOMAIN WITH COVALENTLY LINKED NOVOLACTONE | | Descriptor: | (5beta,6alpha,8alpha,14alpha)-13-ethenyl-5,6-dihydroxy-14-methylpodocarp-12-en-15-oic acid, Heat shock 70 kDa protein 1A/1B | | Authors: | Kirby, C.A, Baird, J, Stams, T. | | Deposit date: | 2014-11-04 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The novolactone natural product disrupts the allosteric regulation of hsp70.
Chem.Biol., 22, 2015
|
|
4WV5
 
 | |
6B0F
 
 | |
1SMK
 
 | | Mature and translocatable forms of glyoxysomal malate dehydrogenase have different activities and stabilities but similar crystal structures | | Descriptor: | CITRIC ACID, Malate dehydrogenase, glyoxysomal | | Authors: | Cox, B, Chit, M.M, Weaver, T, Bailey, J, Gietl, C, Bell, E, Banaszak, L. | | Deposit date: | 2004-03-09 | | Release date: | 2005-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Organelle and translocatable forms of glyoxysomal malate dehydrogenase. The effect of the N-terminal presequence.
Febs J., 272, 2005
|
|
1SEV
 
 | | Mature and translocatable forms of glyoxysomal malate dehydrogenase have different activities and stabilities but similar crystal structures | | Descriptor: | Malate dehydrogenase, glyoxysomal precursor | | Authors: | Cox, B.R, Chit, M.M, Weaver, T.M, Bailey, J, Gietl, C, Bell, E, Banaszak, L.J. | | Deposit date: | 2004-02-18 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Organelle and translocatable forms of glyoxysomal malate dehydrogenase. The effect of the N-terminal presequence
Febs J., 272, 2005
|
|
3JPV
 
 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a pyrrolo[2,3-a]carbazole ligand | | Descriptor: | 1,10-dihydropyrrolo[2,3-a]carbazole-3-carbaldehyde, Peptide (PIMTIDE) ARKRRRHPSGPPTA, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Filippakopoulos, P, Bullock, A.N, Fedorov, O, Akue-Gedu, R, Rossignol, E, Azzaro, S, Bain, J, Cohen, P, Prudhomme, M, Moreau, P, Amizon, F, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-04 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Synthesis, kinase inhibitory potencies, and in vitro antiproliferative evaluation of new pim kinase inhibitors.
J.Med.Chem., 52, 2009
|
|
5N6L
 
 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt C387A mutant from E. coli | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, GLYCEROL | | Authors: | Huang, C.-Y, Boland, C, Howe, N, Wiktor, M, Vogeley, L, Weichert, D, Bailey, J, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis.
Nat Commun, 8, 2017
|
|
5N6H
 
 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, GLYCEROL | | Authors: | Huang, C.-Y, Boland, C, Howe, N, Wiktor, M, Vogeley, L, Weichert, D, Bailey, J, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis.
Nat Commun, 8, 2017
|
|
5N6M
 
 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt from P. aeruginosa | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, CITRATE ANION, ... | | Authors: | Huang, C.-Y, Boland, C, Howe, N, Wiktor, M, Vogeley, L, Weichert, D, Bailey, J, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis.
Nat Commun, 8, 2017
|
|
5T92
 
 | | ESTROGEN RECEPTOR ALPHA LIGAND BINDING DOMAIN IN COMPLEX WITH (2E)-3-{4-[(1R)-2-(4-fluorophenyl)-6-hydroxy-1-methy l-1,2,3,4- tetrahydroisoquinolin-1-yl]phenyl}prop-2-enoic acid | | Descriptor: | (2E)-3-{4-[(1R)-2-(4-fluorophenyl)-6-hydroxy-1-methyl-1,2,3,4-tetrahydroisoquinolin-1-yl]phenyl}prop-2-enoic acid, Estrogen receptor | | Authors: | Kirby, C, Baird, J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery of an Acrylic Acid Based Tetrahydroisoquinoline as an Orally Bioavailable Selective Estrogen Receptor Degrader for ER alpha + Breast Cancer.
J. Med. Chem., 60, 2017
|
|
