3R02
 
 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 7-[(cis-4-aminocyclohexyl)amino]-5-bromo-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Xiang, Y, Hirth, B, Asmussen, G, Biemann, H.-P, Good, A, Fitzgerald, M, Gladysheva, T, Jancsics, K, Liu, J, Metz, M, Papoulis, A, Skerlj, R, Stepp, D.J, Wei, R.R. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3R04
 
 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 5-{6-[(trans-4-aminocyclohexyl)amino]pyrazin-2-yl}-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Xiang, Y, Hirth, B, Asmussen, G, Biemann, H.-P, Good, A, Fitzgerald, M, Gladysheva, T, Jancsics, K, Liu, J, Metz, M, Papoulis, A, Skerlj, R, Stepp, D.J, Wei, R.R. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
8W1L
 
 | |
3R00
 
 | |
3R01
 
 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 5-bromo-7-methoxy-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Liu, J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3VC4
 
 | |
3VBT
 
 | |
3VBV
 
 | |
3VBQ
 
 | |
3VBY
 
 | |
3VBW
 
 | |
3VBX
 
 | |
3ENG
 
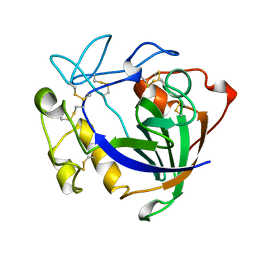 | | STRUCTURE OF ENDOGLUCANASE V CELLOBIOSE COMPLEX | | Descriptor: | ENDOGLUCANASE V CELLOBIOSE COMPLEX, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1996-10-17 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure determination and refinement of the Humicola insolens endoglucanase V at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
4ENG
 
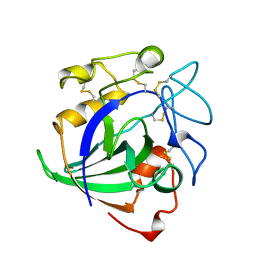 | | STRUCTURE OF ENDOGLUCANASE V CELLOHEXAOSE COMPLEX | | Descriptor: | ENDOGLUCANASE V CELLOHEXAOSE COMPLEX, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1996-10-17 | | Release date: | 1997-06-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure determination and refinement of the Humicola insolens endoglucanase V at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1OVW
 
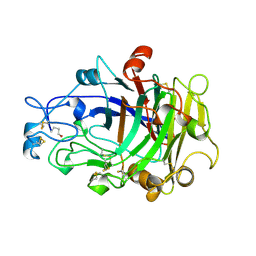 | | ENDOGLUCANASE I COMPLEXED WITH NON-HYDROLYSABLE SUBSTRATE ANALOGUE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-thio-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-1,4-dithio-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Sulzenbacher, G, Davies, G.J, Schulein, M. | | Deposit date: | 1996-10-17 | | Release date: | 1997-10-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Fusarium oxysporum endoglucanase I with a nonhydrolyzable substrate analogue: substrate distortion gives rise to the preferred axial orientation for the leaving group.
Biochemistry, 35, 1996
|
|
2ENG
 
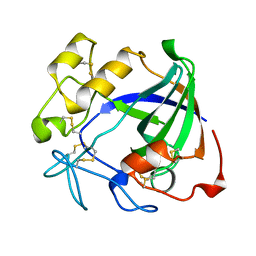 | | ENDOGLUCANASE V | | Descriptor: | ENDOGLUCANASE V | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1995-06-29 | | Release date: | 1996-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of oligosaccharide-bound forms of the endoglucanase V from Humicola insolens at 1.9 A resolution.
Biochemistry, 34, 1995
|
|
