7WT4
 
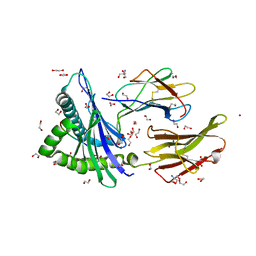 | | Crystal structure of HLA-A*2402 complexed with 8-mer Influenza PB1 peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Asa, M, Morita, D, Sugita, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89459145 Å) | | Cite: | Crystal structures of N-myristoylated lipopeptide-bound HLA class I complexes indicate reorganization of B-pocket architecture upon ligand binding.
J.Biol.Chem., 298, 2022
|
|
7WT5
 
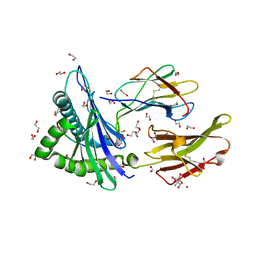 | | Crystal structure of HLA-A*2450 complexed with 8-mer model peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 8-mer model peptide, ... | | Authors: | Asa, M, Morita, D, Sugita, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.0950768 Å) | | Cite: | Crystal structures of N-myristoylated lipopeptide-bound HLA class I complexes indicate reorganization of B-pocket architecture upon ligand binding.
J.Biol.Chem., 298, 2022
|
|
7WT3
 
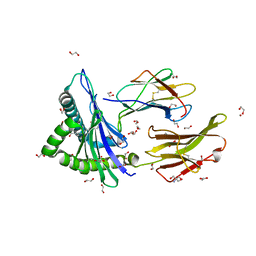 | | Crystal structure of HLA-A*2402 complexed with 4-mer lipopeptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-mer lipopeptide, ... | | Authors: | Asa, M, Morita, D, Sugita, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.887655 Å) | | Cite: | Crystal structures of N-myristoylated lipopeptide-bound HLA class I complexes indicate reorganization of B-pocket architecture upon ligand binding.
J.Biol.Chem., 298, 2022
|
|
6EMK
 
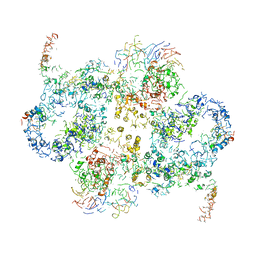 | | Cryo-EM Structure of Saccharomyces cerevisiae Target of Rapamycin Complex 2 | | Descriptor: | Serine/threonine-protein kinase TOR2, Target of rapamycin complex 2 subunit AVO1, Target of rapamycin complex 2 subunit AVO2, ... | | Authors: | Karuppasamy, M, Kusmider, B, Oliveira, T.M, Gaubitz, C, Prouteau, M, Loewith, R, Schaffitzel, C. | | Deposit date: | 2017-10-02 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Cryo-EM structure of Saccharomyces cerevisiae target of rapamycin complex 2.
Nat Commun, 8, 2017
|
|
1EUZ
 
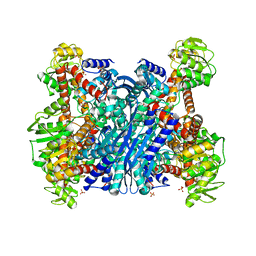 | | GLUTAMATE DEHYDROGENASE FROM THERMOCOCCUS PROFUNDUS IN THE UNLIGATED STATE | | Descriptor: | GLUTAMATE DEHYDROGENASE, SULFATE ION | | Authors: | Nakasako, M. | | Deposit date: | 2000-04-19 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Large-scale domain movements and hydration structure changes in the active-site cleft of unligated glutamate dehydrogenase from Thermococcus profundus studied by cryogenic X-ray crystal structure analysis and small-angle X-ray scattering.
Biochemistry, 40, 2001
|
|
6GRL
 
 | |
8GSU
 
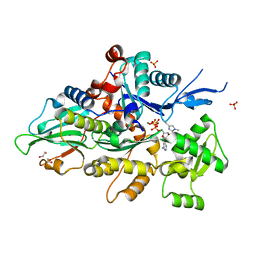 | | Crystal structure of human cardiac alpha actin (WT_ADP-Pi) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
8GT4
 
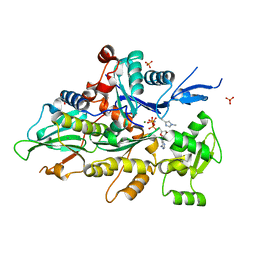 | | Crystal structure of human cardiac alpha actin Q137A mutant (AMPPNP state) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, Actin, alpha cardiac muscle 1, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
8GT1
 
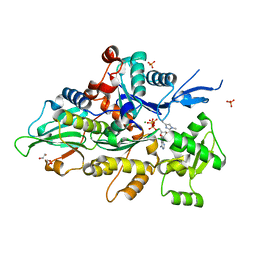 | | Crystal structure of human cardiac alpha actin A108G mutant (ADP-Pi state) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
8GSW
 
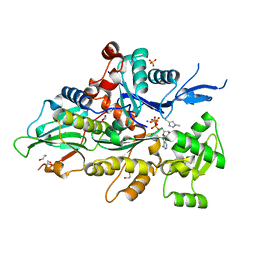 | | Crystal structure of human cardiac alpha actin A108G mutant (AMPPNP state) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, Actin, alpha cardiac muscle 1, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
8GT2
 
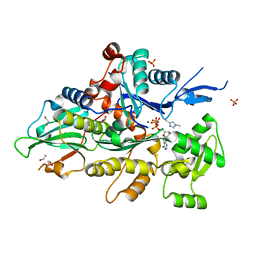 | | Crystal structure of human cardiac alpha actin P109A mutant (AMPPNP state) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, Actin, alpha cardiac muscle 1, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
8GT5
 
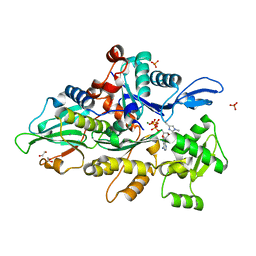 | | Crystal structure of human cardiac alpha actin Q137A mutant (ADP-Pi state) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
8GT3
 
 | | Crystal structure of human cardiac alpha actin P109A mutant (ADP-Pi state) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
6JNR
 
 | | RXRa structure complexed with CU-6PMN and SRC1 peptide. | | Descriptor: | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid, HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Retinoic acid receptor RXR-alpha | | Authors: | Kawasaki, M, Nakano, S, Motoyama, T, Yamada, S, Watanabe, M, Takamura, Y, Fujihara, M, Tokiwa, H, Kakuta, H, Ito, S. | | Deposit date: | 2019-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RXRa structure complexed with CU-6PMN and SRC1 peptide.
To Be Published
|
|
7W97
 
 | | Crystal Structure of the CYP102A1 (P450BM3) Heme Domain with N-Hexadecanoyl-L-Homoserine | | Descriptor: | (2~{S})-2-(hexadecanoylamino)-4-oxidanyl-butanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Karasawa, M, Stanfield, J.K, Kasai, C, Sugimoto, H, Shoji, O. | | Deposit date: | 2021-12-09 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the CYP102A1 (P450BM3) Heme Domain with N-Hexadecanoyl-L-Homoserine
To Be Published
|
|
7W9D
 
 | | Crystal Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 (P450BM3) Heme Domain with N-Hexadecanoyl-L-Homoserine | | Descriptor: | (2~{S})-2-(hexadecanoylamino)-4-oxidanyl-butanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Karasawa, M, Stanfield, J.K, Kasai, C, Sugimoto, H, Shoji, O. | | Deposit date: | 2021-12-09 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 (P450BM3) Heme Domain with N-Hexadecanoyl-L-Homoserine
To Be Published
|
|
7W9J
 
 | | Crystal Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 (P450BM3) Heme Domain with N-Dodecanoyl-L-Homoserine Lactone | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, N-[(3S)-2-oxooxolan-3-yl]dodecanamide, ... | | Authors: | Karasawa, M, Stanfield, J.K, Kasai, C, Sugimoto, H, Shoji, O. | | Deposit date: | 2021-12-09 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 (P450BM3) Heme Domain with N-Dodecanoyl-L-Homoserine Lactone
To Be Published
|
|
5B6Q
 
 | | Crystal structure of monomeric cytochrome c5 from Shewanella violacea | | Descriptor: | HEME C, IMIDAZOLE, Soluble cytochrome cA | | Authors: | Masanari, M, Fujii, S, Kawahara, K, Oki, H, Tsujino, H, Maruno, T, Kobayashi, Y, Ohkubo, T, Nishiyama, M, Harada, Y, Wakai, S, Sambongi, Y. | | Deposit date: | 2016-06-01 | | Release date: | 2016-10-19 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Comparative study on stabilization mechanism of monomeric cytochrome c5 from deep-sea piezophilic Shewanella violacea
Biosci.Biotechnol.Biochem., 2016
|
|
6HZN
 
 | | Crystal structure of human dermatan sulfate epimerase 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hasan, M, Unge, J, Westergren-Thorsson, G, Ellervik, U, Mueller, U, Malmstrom, A, Tykesson, E. | | Deposit date: | 2018-10-23 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The structure of human dermatan sulfate epimerase 1 emphasizes the importance of C5-epimerization of glucuronic acid in higher organisms
Chem Sci, 2020
|
|
6L96
 
 | | Structure of PPARalpha-LBD/pemafibrate/SRC1 peptide | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, Peroxisome proliferator-activated receptor alpha, SRC1 coactivator peptide | | Authors: | Kawasaki, M, Kambe, A, Yamamoto, Y, Arulmozhira, S, Ito, S, Nakagawa, Y, Tokiwa, H, Nakano, S, Shimano, H. | | Deposit date: | 2019-11-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Elucidation of Molecular Mechanism of a Selective PPAR alpha Modulator, Pemafibrate, through Combinational Approaches of X-ray Crystallography, Thermodynamic Analysis, and First-Principle Calculations.
Int J Mol Sci, 21, 2020
|
|
2Z6D
 
 | |
2Z6C
 
 | |
1DLF
 
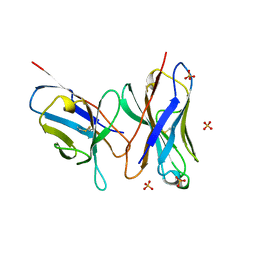 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 5.25 | | Descriptor: | ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S), SULFATE ION | | Authors: | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | Deposit date: | 1998-07-14 | | Release date: | 1999-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
1IO6
 
 | |
1JTI
 
 | |
