7MEQ
 
 | | Crystal structure of human TMPRSS2 in complex with Nafamostat | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, Transmembrane protease serine 2, ... | | Authors: | Fraser, B, Beldar, S, Hutchinson, A, Li, Y, Seitova, A, Edwards, A.M, Benard, F, Arrowsmith, C.H, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-07 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and activity of human TMPRSS2 protease implicated in SARS-CoV-2 activation.
Nat.Chem.Biol., 18, 2022
|
|
7A21
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2158 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(pyrrolidin-1-ylmethyl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type I, ... | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-08-14 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2153
To Be Published
|
|
6ZU8
 
 | | Crystal structure of human Brachyury G177D variant in complex with Afatinib | | Descriptor: | Brachyury protein, N-{4-[(3-chloro-4-fluorophenyl)amino]-7-[(3S)-tetrahydrofuran-3-yloxy]quinazolin-6-yl}-4-(dimethylamino)butanamide, ZINC ION | | Authors: | Newman, J.A, Gavard, A.E, Shrestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-07-21 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human Brachyury G177D variant in complex with Afatinib
To Be Published
|
|
1PU1
 
 | | Solution structure of the Hypothetical protein mth677 from Methanothermobacter Thermautotrophicus | | Descriptor: | Hypothetical protein MTH677 | | Authors: | Blanco, F.J, Yee, A, Campos-Olivas, R, Devos, D, Valencia, A, Arrowsmith, C.H, Rico, M. | | Deposit date: | 2003-06-23 | | Release date: | 2004-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the hypothetical protein Mth677 from Methanobacterium thermoautotrophicum: A novel {alpha}+{beta} fold
Protein Sci., 13, 2004
|
|
1HS5
 
 | | NMR SOLUTION STRUCTURE OF DESIGNED P53 DIMER | | Descriptor: | CELLULAR TUMOR ANTIGEN P53 | | Authors: | Davison, T.S, Nie, X, Ma, W, Li, Y, Kay, C, Benchimol, S, Arrowsmith, C.H. | | Deposit date: | 2000-12-22 | | Release date: | 2001-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and functionality of a designed p53 dimer.
J.Mol.Biol., 307, 2001
|
|
1HYW
 
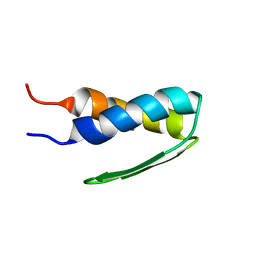 | | SOLUTION STRUCTURE OF BACTERIOPHAGE LAMBDA GPW | | Descriptor: | HEAD-TO-TAIL JOINING PROTEIN W | | Authors: | Maxwell, K.L, Yee, A.A, Booth, V, Arrowsmith, C.H, Gold, M, Davidson, A.R. | | Deposit date: | 2001-01-22 | | Release date: | 2001-04-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of bacteriophage lambda protein W, a small morphogenetic protein possessing a novel fold.
J.Mol.Biol., 308, 2001
|
|
1PET
 
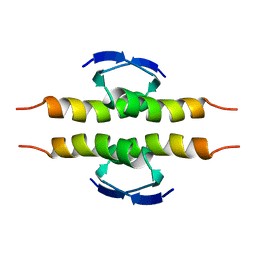 | | NMR SOLUTION STRUCTURE OF THE TETRAMERIC MINIMUM TRANSFORMING DOMAIN OF P53 | | Descriptor: | TUMOR SUPPRESSOR P53 | | Authors: | Lee, W, Harvey, T.S, Yin, Y, Yau, P, Litchfield, D, Arrowsmith, C.H. | | Deposit date: | 1994-11-24 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tetrameric minimum transforming domain of p53.
Nat.Struct.Biol., 1, 1994
|
|
4UY4
 
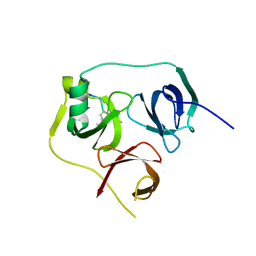 | | 1.86 A structure of human Spindlin-4 protein in complex with histone H3K4me3 peptide | | Descriptor: | GLYCEROL, HISTONE H3K4ME3, SPINDLIN-4 | | Authors: | Talon, R, Gileadi, C, Johansson, C, Burgess-Brown, N, Shrestha, L, von Delft, F, Krojer, T, Fairhead, M, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U. | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | 1.86 A Structure of Human Spindlin-4 Protein in Complex with Histone H3K4Me3 Peptide
To be Published
|
|
6OOV
 
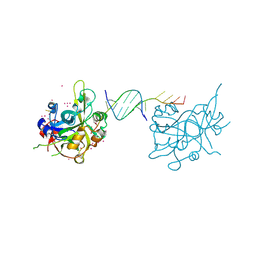 | | Crystal structure of HMCES SRAP domain in complex with palindromic 3' overhang DNA | | Descriptor: | DNA (5'-D(*CP*AP*AP*CP*GP*TP*TP*GP*TP*TP*TP*TP*T)-3'), Embryonic stem cell-specific 5-hydroxymethylcytosine-binding protein, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Zeng, H, Li, Y, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-23 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of HMCES SRAP domain in complex with palindromic 3' overhang DNA
To Be Published
|
|
6OEA
 
 | | Crystal structure of HMCES SRAP domain in complex with longer 3' overhang DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*AP*CP*GP*TP*TP*GP*TP*T)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*G)-3'), ... | | Authors: | Halabelian, L, Ravichandran, M, Li, Y, Zeng, H, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OE7
 
 | | Crystal structure of HMCES cross-linked to DNA abasic site | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*AP*CP*GP*TP*(DRZ)P*GP*TP*T)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*G)-3'), ... | | Authors: | Halabelian, L, Li, Y, Zeng, H, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OQM
 
 | | crystal structure of the MSH6 PWWP domain | | Descriptor: | DNA mismatch repair protein Msh6, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-26 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | crystal structure of the MSH6 PWWP domain
To Be Published
|
|
7RBQ
 
 | | Co-crystal structure of human PRMT9 in complex with MT556 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 7-[5-S-(4-{[(4-ethylpyridin-3-yl)methyl]amino}butyl)-5-thio-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Protein arginine N-methyltransferase 9, ... | | Authors: | Zeng, H, Dong, A, Hutchinson, A, Seitova, A, Li, Y, Gao, Y.D, Schneider, S, Siliphaivanh, P, Sloman, D, Nicholson, B, Fischer, C, Hicks, J, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-06 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Co-crystal structure of human PRMT9 in complex with MT556 inhibitor
To Be Published
|
|
7R4N
 
 | | Structure of human hydroxyacid oxidase 1 bound with 5-bromo-N-methyl-1H-indazole-3-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-N-methyl-1H-indazole-3-carboxamide, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Mackinnon, S, Bezerra, G.A, Krojer, T, Bradley, A.R, Talon, R, Brandeo-Neto, J, Douangamath, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W. | | Deposit date: | 2022-02-08 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human hydroxyacid oxidase 1 bound with 5-bromo-N-methyl-1H-indazole-3-carboxamide
To Be Published
|
|
7R4O
 
 | | Structure of human hydroxyacid oxidase 1 bound with 2-((4H-1,2,4-triazol-3-yl)thio)-1-(4-(3-chlorophenyl)piperazin-1-yl)ethan-1-one | | Descriptor: | 2-((4H-1,2,4-triazol-3-yl)thio)-1-(4-(3-chlorophenyl)piperazin-1-yl)ethan-1-one, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Mackinnon, S, Bezerra, G.A, Krojer, T, Bradley, A.R, Talon, R, Brandeo-Neto, J, Douangamath, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W. | | Deposit date: | 2022-02-08 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of human hydroxyacid oxidase 1 bound with 5-bromo-N-methyl-1H-indazole-3-carboxamide
To Be Published
|
|
7R4P
 
 | | Structure of human hydroxyacid oxidase 1 bound with 6-amino-1-benzyl-5-(methylamino)pyrimidine-2,4(1H,3H)-dione | | Descriptor: | 6-amino-1-benzyl-5-(methylamino)pyrimidine-2,4(1H,3H)-dione, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Mackinnon, S, Bezerra, G.A, Krojer, T, Bradley, A.R, Talon, R, Brandeo-Neto, J, Douangamath, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W. | | Deposit date: | 2022-02-09 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure of human hydroxyacid oxidase 1 bound with 5-bromo-N-methyl-1H-indazole-3-carboxamide
To Be Published
|
|
8W3V
 
 | | Crystal structure of human WDR41 | | Descriptor: | WD repeat-containing protein 41 | | Authors: | Hutchinson, A, Dong, A, Li, Y, Seitova, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-02-22 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human WDR41
To be published
|
|
4DIP
 
 | | Crystal structure of human Peptidyl-prolyl cis-trans isomerase FKBP14 | | Descriptor: | PHOSPHATE ION, Peptidyl-prolyl cis-trans isomerase FKBP14, SODIUM ION | | Authors: | Krojer, T, Kiyani, W, Goubin, S, Muniz, J.R.C, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Oppermann, U, Zschocke, J, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-01-31 | | Release date: | 2012-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of human Peptidyl-prolyl cis-trans isomerase FKBP14
To be Published
|
|
7M3X
 
 | | Crystal Structure of the Apo Form of Human RBBP7 | | Descriptor: | Histone-binding protein RBBP7, UNKNOWN ATOM OR ION | | Authors: | Righetto, G.L, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-19 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of the Apo Form of Human RBBP7
To Be Published
|
|
7MDN
 
 | | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound MRT10241866a | | Descriptor: | Histone-lysine N-methyltransferase NSD2, UNKNOWN ATOM OR ION, ~{N}-cyclopropyl-3-oxidanylidene-~{N}-(thiophen-2-ylmethyl)-4~{H}-1,4-benzoxazine-7-carboxamide | | Authors: | Lei, M, Freitas, R.F, Dong, A, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-05 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization.
Nat.Chem.Biol., 18, 2022
|
|
7M40
 
 | | Discovery of small molecule antagonists of human Retinoblastoma Binding Protein 4 (RBBP4) | | Descriptor: | Histone-binding protein RBBP4, N~3~-{4-[3-(dimethylamino)pyrrolidin-1-yl]-6,7-dimethoxyquinazolin-2-yl}-N~1~,N~1~-dimethylpropane-1,3-diamine | | Authors: | Perveen, S, Dong, A, Tempel, W, Zepeda-Velazquez, C, Abbey, M, McLeod, D, Marcellus, R, Mohammed, M, Ensan, D, Panagopoulos, D, Trush, V, Gibson, E, Brown, P.J, Arrowsmith, C.H, Schapira, M, Al-awar, R, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-19 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of small molecule antagonists of human Retinoblastoma Binding Protein 4 (RBBP4)
To Be Published
|
|
4DYM
 
 | | Crystal structure of the ACVR1 kinase domain in complex with the imidazo[1,2-b]pyridazine inhibitor K00135 | | Descriptor: | 1-(3-{6-[(CYCLOPROPYLMETHYL)AMINO]IMIDAZO[1,2-B]PYRIDAZIN-3-YL}PHENYL)ETHANONE, Activin receptor type-1, GLYCEROL, ... | | Authors: | Chaikuad, A, Sanvitale, C, Cooper, C, Canning, P, Mahajan, P, Daga, N, Petrie, K, Alfano, I, Gileadi, O, Fedorov, O, Krojer, T, Filippakopoulos, P, Muniz, J.R.C, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of the ACVR1 kinase domain in complex with the imidazo[1,2-b]pyridazine inhibitor K00135
To be Published
|
|
4DYL
 
 | | F-BAR domain of human FES tyrosine kinase | | Descriptor: | Tyrosine-protein kinase Fes/Fps | | Authors: | Ugochukwu, E, Salah, E, Elkins, J, Barr, A, Krojer, T, Filippakopoulos, P, Weigelt, J, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-02-29 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | F-BAR domain of human FES tyrosine kinase
TO BE PUBLISHED
|
|
4ER5
 
 | | Crystal structure of human DOT1L in complex with 2 molecules of EPZ004777 | | Descriptor: | 7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Wernimont, A.K, Tempel, W, Yu, W, Scopton, A, Li, Y, Nguyen, K.T, Federation, A, Marineau, J, Qi, J, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
4E71
 
 | | Crystal structure of the RHO GTPASE binding domain of Plexin B2 | | Descriptor: | Plexin-B2, SODIUM ION | | Authors: | Guan, X, Wang, H, Tempel, W, Tong, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-16 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of the RHO GTPASE binding domain of Plexin B2
to be published
|
|
