2JEX
 
 | | Transcription activator structure reveals redox control of a replication initiation reaction | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Sanders, C.M, Sizov, D, Seavers, P.R, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2007-01-24 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Transcription Activator Structure Reveals Redox Control of a Replication Initiation Reaction.
Nucleic Acids Res., 35, 2007
|
|
2JK3
 
 | |
2JEU
 
 | | Transcription activator structure reveals redox control of a replication initiation reaction | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Sanders, C.M, Sizov, D, Seavers, P.R, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2007-01-23 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Transcription Activator Structure Reveals Redox Control of a Replication Initiation Reaction.
Nucleic Acids Res., 35, 2007
|
|
3ZQO
 
 | | Crystal structure of the small terminase oligomerization core domain from a SPP1-like bacteriophage (crystal form 3) | | Descriptor: | POTASSIUM ION, TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZZL
 
 | | Bacillus halodurans trp RNA-binding attenuation protein (TRAP): a 12- subunit assembly | | Descriptor: | TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Chen, C, Smits, C, Dodson, G.G, Shevtsov, M.B, Merlino, N, Gollnick, P, Antson, A.A. | | Deposit date: | 2011-09-01 | | Release date: | 2011-10-12 | | Last modified: | 2011-10-26 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | How to Change the Oligomeric State of a Circular Protein Assembly: Switch from 11-Subunit to 12-Subunit Trap Suggests a General Mechanism
Plos One, 6, 2011
|
|
3ZQP
 
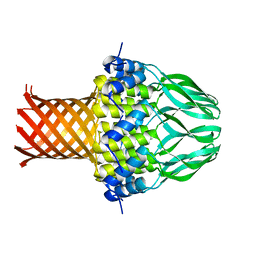 | | Crystal structure of the small terminase oligomerization domain from a SPP1-like bacteriophage | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZZQ
 
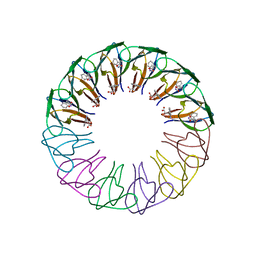 | | Engineered 12-subunit Bacillus subtilis trp RNA-binding attenuation protein (TRAP) | | Descriptor: | TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Chen, C, Smits, C, Dodson, G.G, Shevtsov, M.B, Merlino, N, Gollnick, P, Antson, A.A. | | Deposit date: | 2011-09-02 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | How to Change the Oligomeric State of a Circular Protein Assembly: Switch from 11-Subunit to 12-Subunit Trap Suggests a General Mechanism
Plos One, 6, 2011
|
|
3ZZS
 
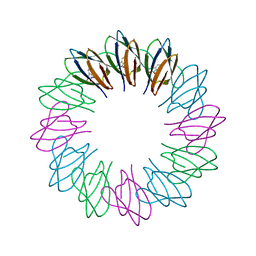 | | Engineered 12-subunit Bacillus stearothermophilus trp RNA-binding attenuation protein (TRAP) | | Descriptor: | TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Chen, C, Smits, C, Dodson, G.G, Shevtsov, M.B, Merlino, N, Gollnick, P, Antson, A.A. | | Deposit date: | 2011-09-02 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | How to Change the Oligomeric State of a Circular Protein Assembly: Switch from 11-Subunit to 12-Subunit Trap Suggests a General Mechanism
Plos One, 6, 2011
|
|
4B27
 
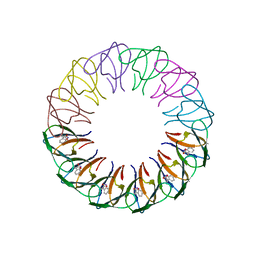 | | Trp RNA-binding attenuation protein: modifying symmetry and stability of a circular oligomer | | Descriptor: | TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Bayfield, O.W, Chen, C, Patterson, A.R, Luan, W, Smits, C, Gollnick, P, Antson, A.A. | | Deposit date: | 2012-07-12 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Trp RNA-Binding Attenuation Protein: Modifying Symmetry and Stability of a Circular Oligomer.
Plos One, 7, 2012
|
|
4BF9
 
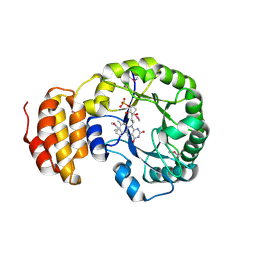 | | Crystal structure of E. coli dihydrouridine synthase C (DusC) (selenomethionine derivative) | | Descriptor: | FLAVIN MONONUCLEOTIDE, TRNA-DIHYDROURIDINE SYNTHASE C | | Authors: | Byrne, R.T, Whelan, F, Konevega, A, Aziz, N, Rodnina, M, Antson, A.A. | | Deposit date: | 2013-03-16 | | Release date: | 2013-11-06 | | Last modified: | 2015-05-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Major Reorientation of tRNA Substrates Defines Specificity of Dihydrouridine Synthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4BFA
 
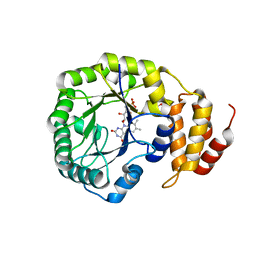 | | Crystal structure of E. coli dihydrouridine synthase C (DusC) | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, NITRATE ION, ... | | Authors: | Byrne, R.T, Whelan, F, Konevega, A, Aziz, N, Rodnina, M, Antson, A.A. | | Deposit date: | 2013-03-16 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Major Reorientation of tRNA Substrates Defines Specificity of Dihydrouridine Synthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2VLH
 
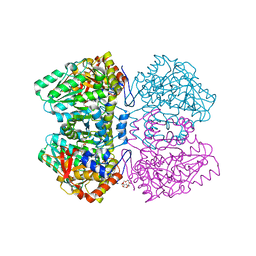 | | Quinonoid intermediate of Citrobacter freundii tyrosine phenol-lyase formed with methionine | | Descriptor: | (2E)-2-{[(Z)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4(1H)-YLIDENE}METHYL]IMINO}-4-(METHYLSULFANYL)BUTANOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | Authors: | Milic, D, Demidkina, T.V, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2008-01-14 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights Into the Catalytic Mechanism of Tyrosine Phenol-Lyase from X-Ray Structures of Quinonoid Intermediates.
J.Biol.Chem., 283, 2008
|
|
2VLF
 
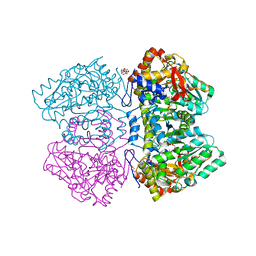 | | Quinonoid intermediate of Citrobacter freundii tyrosine phenol-lyase formed with alanine | | Descriptor: | (2E)-2-{[(Z)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4(1H)-YLIDENE}METHYL]IMINO}PROPANOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | Authors: | Milic, D, Demidkina, T.V, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2008-01-14 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Insights Into the Catalytic Mechanism of Tyrosine Phenol-Lyase from X-Ray Structures of Quinonoid Intermediates.
J.Biol.Chem., 283, 2008
|
|
2WBN
 
 | | Crystal structure of the g2p (large terminase) nuclease domain from the bacteriophage SPP1 | | Descriptor: | TERMINASE LARGE SUBUNIT | | Authors: | Smits, C, Chechik, M, Kovalevskiy, O.V, Shevtsov, M.B, Foster, A.W, Alonso, J.C, Antson, A.A. | | Deposit date: | 2009-03-02 | | Release date: | 2009-03-24 | | Last modified: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Nuclease Activity of a Bacteriophage Large Terminase.
Embo Rep., 10, 2009
|
|
2WC9
 
 | | Crystal structure of the g2p (large terminase) nuclease domain from the bacteriophage SPP1 with bound Mn | | Descriptor: | MANGANESE (II) ION, SULFATE ION, TERMINASE LARGE SUBUNIT | | Authors: | Smits, C, Chechik, M, Kovalevskiy, O.V, Shevtsov, M.B, Foster, A.W, Alonso, J.C, Antson, A.A. | | Deposit date: | 2009-03-10 | | Release date: | 2009-03-24 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Nuclease Activity of a Bacteriophage Large Terminase.
Embo Rep., 10, 2009
|
|
2WNR
 
 | | The structure of Methanothermobacter thermautotrophicus exosome core assembly | | Descriptor: | PHOSPHATE ION, PROBABLE EXOSOME COMPLEX EXONUCLEASE 1, PROBABLE EXOSOME COMPLEX EXONUCLEASE 2 | | Authors: | Ng, C.L, Waterman, D.G, Antson, A.A, Ortiz-Lombardia, M. | | Deposit date: | 2009-07-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the Methanothermobacter Thermautotrophicus Exosome Rnase Ph Ring
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2WV1
 
 | |
2YCB
 
 | | Structure of the archaeal beta-CASP protein with N-terminal KH domains from Methanothermobacter thermautotrophicus | | Descriptor: | CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Silva, A.P.G, Chechik, M, Byrne, R.T, Waterman, D.G, Ng, C.L, Dodson, E.J, Koonin, E.V, Antson, A.A, Smits, C. | | Deposit date: | 2011-03-13 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and Activity of a Novel Archaeal Beta-Casp Protein with N-Terminal Kh Domains.
Structure, 19, 2011
|
|
2YCP
 
 | | F448H mutant of tyrosine phenol-lyase from Citrobacter freundii in complex with quinonoid intermediate formed with 3-fluoro-L-tyrosine | | Descriptor: | (2E)-3-(3-fluoro-4-hydroxyphenyl)-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}propanoic acid, 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Milic, D, Demidkina, T.V, Faleev, N.G, Phillips, R.S, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2011-03-16 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Snapshots of Tyrosine Phenol-Lyase Show that Substrate Strain Plays a Role in C-C Bond Cleavage
J.Am.Chem.Soc., 133, 2011
|
|
2YHK
 
 | | D214A mutant of tyrosine phenol-lyase from Citrobacter freundii | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, DI(HYDROXYETHYL)ETHER, POTASSIUM ION, ... | | Authors: | Milic, D, Demidkina, T.V, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2011-05-03 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal Structure of Citrobacter Freundii Asp214Ala Tyrosine Phenol-Lyase Reveals that Asp214 is Critical for Maintaining a Strain in the Internal Aldimine
Croatica Chemica Acta, 85, 2012
|
|
2YCT
 
 | | Tyrosine phenol-lyase from Citrobacter freundii in complex with pyridine N-oxide and the quinonoid intermediate formed with L-alanine | | Descriptor: | (2E)-2-{[(Z)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4(1H)-YLIDENE}METHYL]IMINO}PROPANOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, PHOSPHATE ION, ... | | Authors: | Milic, D, Demidkina, T.V, Faleev, N.G, Phillips, R.S, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2011-03-16 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic Snapshots of Tyrosine Phenol-Lyase Show that Substrate Strain Plays a Role in C-C Bond Cleavage
J.Am.Chem.Soc., 133, 2011
|
|
2YCN
 
 | | Y71F mutant of tyrosine phenol-lyase from Citrobacter freundii in complex with quinonoid intermediate formed with 3-fluoro-L-tyrosine | | Descriptor: | (2E)-3-(3-fluoro-4-hydroxyphenyl)-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}propanoic acid, DI(HYDROXYETHYL)ETHER, POTASSIUM ION, ... | | Authors: | Milic, D, Demidkina, T.V, Faleev, N.G, Phillips, R.S, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2011-03-16 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystallographic Snapshots of Tyrosine Phenol-Lyase Show that Substrate Strain Plays a Role in C-C Bond Cleavage
J.Am.Chem.Soc., 133, 2011
|
|
1TPL
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF TYROSINE PHENOL-LYASE | | Descriptor: | SULFATE ION, TYROSINE PHENOL-LYASE | | Authors: | Antson, A, Demidkina, T, Dauter, Z, Harutyunyan, E, Wilson, K. | | Deposit date: | 1992-11-25 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of tyrosine phenol-lyase.
Biochemistry, 32, 1993
|
|
5W19
 
 | | Tryptophan indole-lyase complex with oxindolyl-L-alanine | | Descriptor: | 1-carboxy-1-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]azaniumyl}-2-[(3R)-2-oxo-2,3-dihydro-1H-indol-3-yl]ethan-1-ide, POTASSIUM ION, Tryptophanase | | Authors: | Phillips, R.S, Wood, Z.A. | | Deposit date: | 2017-06-02 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of Proteus vulgaris tryptophan indole-lyase complexed with oxindolyl-L-alanine: implications for the reaction mechanism.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
2RBI
 
 | | STRUCTURE OF BINASE MUTANT HIS 101 ASN | | Descriptor: | RIBONUCLEASE | | Authors: | Offen, W.A, Okorokov, A.L. | | Deposit date: | 1996-11-12 | | Release date: | 1997-03-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RNA cleavage without hydrolysis. Splitting the catalytic activities of binase with Asn101 and Thr101 mutations.
Protein Eng., 10, 1997
|
|
