1WAP
 
 | |
1DTO
 
 | | CRYSTAL STRUCTURE OF THE COMPLETE TRANSACTIVATION DOMAIN OF E2 PROTEIN FROM THE HUMAN PAPILLOMAVIRUS TYPE 16 | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Antson, A.A, Burns, J.E, Moroz, O.V, Scott, D.J, Sanders, C.M, Bronstein, I.B, Dodson, G.G, Wilson, K.S, Maitland, N. | | Deposit date: | 2000-01-13 | | Release date: | 2000-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the intact transactivation domain of the human papillomavirus E2 protein.
Nature, 403, 2000
|
|
1C9S
 
 | | CRYSTAL STRUCTURE OF A COMPLEX OF TRP RNA-BINDING ATTENUATION PROTEIN WITH A 53-BASE SINGLE STRANDED RNA CONTAINING ELEVEN GAG TRIPLETS SEPARATED BY AU DINUCLEOTIDES | | Descriptor: | SINGLE STRANDED RNA (55-MER), TRP RNA-BINDING ATTENUATION PROTEIN, TRYPTOPHAN | | Authors: | Antson, A.A, Dodson, E.J, Dodson, G.G, Greaves, R.B, Chen, X.-P, Gollnick, P. | | Deposit date: | 1999-08-03 | | Release date: | 1999-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA.
Nature, 401, 1999
|
|
1GZI
 
 | | CRYSTAL STRUCTURE OF TYPE III ANTIFREEZE PROTEIN FROM OCEAN POUT, AT 1.8 ANGSTROM RESOLUTION | | Descriptor: | HPLC-12 TYPE III ANTIFREEZE PROTEIN | | Authors: | Antson, A.A, Lewis, S, Roper, D.I, Smith, D.J, Hubbard, R.E. | | Deposit date: | 1996-10-25 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Type III Antifreeze Protein from Ocean Pout
To be Published
|
|
1HG7
 
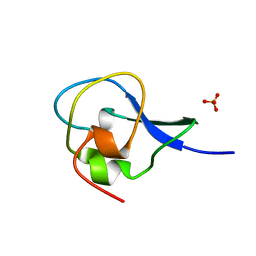 | | High resolution structure of HPLC-12 type III antifreeze protein from Ocean Pout Macrozoarces americanus | | Descriptor: | HPLC-12 TYPE III ANTIFREEZE PROTEIN, SULFATE ION | | Authors: | Antson, A.A, Smith, D.J, Roper, D.I, Lewis, S, Caves, L.S.D, Verma, C.S, Buckley, S.L, Lillford, P.J, Hubbard, R.E. | | Deposit date: | 2000-12-13 | | Release date: | 2001-01-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Understanding the Mechanism of Ice Binding by Type III Antifreeze Protein
J.Mol.Biol., 305, 2001
|
|
4XVN
 
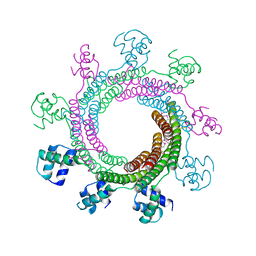 | |
4ZC3
 
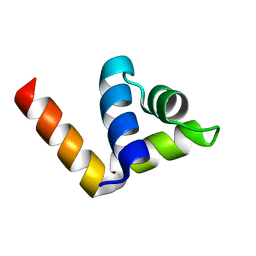 | |
1BYG
 
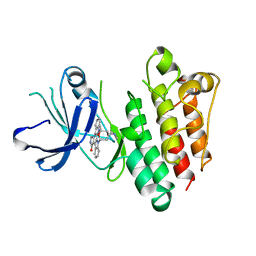 | | KINASE DOMAIN OF HUMAN C-TERMINAL SRC KINASE (CSK) IN COMPLEX WITH INHIBITOR STAUROSPORINE | | Descriptor: | PROTEIN (C-TERMINAL SRC KINASE), STAUROSPORINE | | Authors: | Antson, A.A, Lamers, M.B.A.C, Scott, R.K, Williams, D.H, Hubbard, R.E. | | Deposit date: | 1998-10-14 | | Release date: | 1999-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the protein tyrosine kinase domain of C-terminal Src kinase (CSK) in complex with staurosporine.
J.Mol.Biol., 285, 1999
|
|
2TPL
 
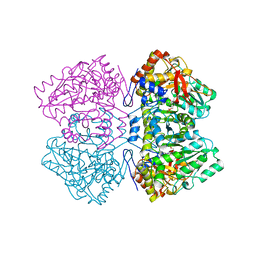 | | TYROSINE PHENOL-LYASE FROM CITROBACTER INTERMEDIUS COMPLEX WITH 3-(4'-HYDROXYPHENYL)PROPIONIC ACID, PYRIDOXAL-5'-PHOSPHATE AND CS+ ION | | Descriptor: | CESIUM ION, HYDROXYPHENYL PROPIONIC ACID, TYROSINE PHENOL-LYASE | | Authors: | Antson, A.A, Demidkina, T.V, Wilson, K.S. | | Deposit date: | 1997-01-23 | | Release date: | 1997-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of Citrobacter freundii tyrosine phenol-lyase complexed with 3-(4'-hydroxyphenyl)propionic acid, together with site-directed mutagenesis and kinetic analysis, demonstrates that arginine 381 is required for substrate specificity.
Biochemistry, 36, 1997
|
|
4V4F
 
 | | The structure of the trp RNA-binding attenuation protein (TRAP) bound to a RNA molecule containing UAGAU repeats | | Descriptor: | 5'-R(*UP*AP*GP*AP*UP)-3', TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Hopcroft, N.H, Manfredo, A, Wendt, A.L, Brzozowski, A.M, Gollnick, P, Antson, A.A. | | Deposit date: | 2003-12-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Interaction of RNA with Trap: The Role of Triplet Repeats and Separating Spacer Nucleotides
J.Mol.Biol., 338, 2004
|
|
2WNY
 
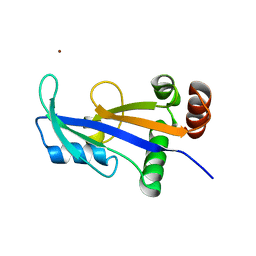 | | Structure of Mth689, a DUF54 protein from Methanothermobacter thermautotrophicus | | Descriptor: | CHLORIDE ION, CONSERVED PROTEIN MTH689, NICKEL (II) ION | | Authors: | Ng, C.L, Waterman, D.G, Lebedev, A.A, Smits, C, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2009-07-21 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Mth689, a Duf54 Protein from Methanothermobacter Thermautotrophicus
To be Published
|
|
8POP
 
 | | HK97 small terminase in complex with DNA | | Descriptor: | DNA (31-MER), Terminase small subunit | | Authors: | Chechik, M, Greive, S.J, Antson, A.A, Jenkins, H.T. | | Deposit date: | 2023-07-05 | | Release date: | 2023-08-09 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for DNA recognition by a viral genome-packaging machine.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2JES
 
 | | Portal protein (gp6) from bacteriophage SPP1 | | Descriptor: | CALCIUM ION, MERCURY (II) ION, PORTAL PROTEIN, ... | | Authors: | Lebedev, A.A, Krause, M.H, Isidro, A.L, Vagin, A.A, Orlova, E.V, Turner, J, Dodson, E.J, Tavares, P, Antson, A.A. | | Deposit date: | 2007-01-21 | | Release date: | 2007-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Framework for DNA Translocation Via the Viral Portal Protein
Embo J., 26, 2007
|
|
2WNR
 
 | | The structure of Methanothermobacter thermautotrophicus exosome core assembly | | Descriptor: | PHOSPHATE ION, PROBABLE EXOSOME COMPLEX EXONUCLEASE 1, PROBABLE EXOSOME COMPLEX EXONUCLEASE 2 | | Authors: | Ng, C.L, Waterman, D.G, Antson, A.A, Ortiz-Lombardia, M. | | Deposit date: | 2009-07-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the Methanothermobacter Thermautotrophicus Exosome Rnase Ph Ring
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4YCP
 
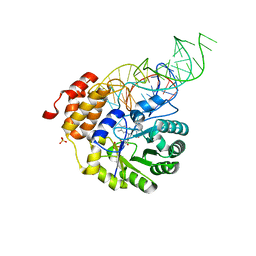 | | E. coli dihydrouridine synthase C (DusC) in complex with tRNATrp | | Descriptor: | FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Byrne, R.T, Jenkins, H.T, Peters, D.T, Whelan, F, Stowell, J, Aziz, N, Kasatsky, P, Rodnina, M.V, Koonin, E.V, Konevega, A.L, Antson, A.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Major reorientation of tRNA substrates defines specificity of dihydrouridine synthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YCO
 
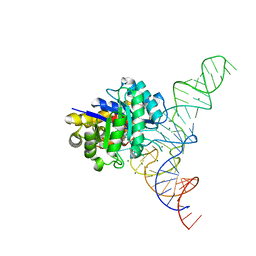 | | E. coli dihydrouridine synthase C (DusC) in complex with tRNAPhe | | Descriptor: | FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Byrne, R.T, Jenkins, H.T, Peters, D.T, Whelan, F, Stowell, J, Aziz, N, Kasatsky, P, Rodnina, M.V, Koonin, E.V, Konevega, A.L, Antson, A.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Major reorientation of tRNA substrates defines specificity of dihydrouridine synthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1ZEG
 
 | | STRUCTURE OF B28 ASP INSULIN IN COMPLEX WITH PHENOL | | Descriptor: | CHLORIDE ION, INSULIN, PHENOL, ... | | Authors: | Whittingham, J.L, Edwards, E.J, Antson, A.A, Clarkson, J.M, Dodson, G.G. | | Deposit date: | 1998-05-01 | | Release date: | 1998-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interactions of phenol and m-cresol in the insulin hexamer, and their effect on the association properties of B28 pro --> Asp insulin analogues.
Biochemistry, 37, 1998
|
|
1ZEH
 
 | | STRUCTURE OF INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, M-CRESOL, ... | | Authors: | Whittingham, J.L, Edwards, E.J, Antson, A.A, Clarkson, J.M, Dodson, G.G. | | Deposit date: | 1998-05-01 | | Release date: | 1998-12-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interactions of phenol and m-cresol in the insulin hexamer, and their effect on the association properties of B28 pro --> Asp insulin analogues.
Biochemistry, 37, 1998
|
|
2JEX
 
 | | Transcription activator structure reveals redox control of a replication initiation reaction | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Sanders, C.M, Sizov, D, Seavers, P.R, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2007-01-24 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Transcription Activator Structure Reveals Redox Control of a Replication Initiation Reaction.
Nucleic Acids Res., 35, 2007
|
|
2JK3
 
 | |
2JEU
 
 | | Transcription activator structure reveals redox control of a replication initiation reaction | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Sanders, C.M, Sizov, D, Seavers, P.R, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2007-01-23 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Transcription Activator Structure Reveals Redox Control of a Replication Initiation Reaction.
Nucleic Acids Res., 35, 2007
|
|
6QJT
 
 | |
7QIK
 
 | | SARS-CoV-2 Nucleocapsid phosphopeptide 193-200 bound to human 14-3-3 sigma | | Descriptor: | 14-3-3 protein sigma, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BROMIDE ION, ... | | Authors: | Sluchanko, N.N, Tugaeva, K.V, Smith, J.L.R, Antson, A.A. | | Deposit date: | 2021-12-15 | | Release date: | 2021-12-29 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Human 14-3-3 Proteins Site-selectively Bind the Mutational Hotspot Region of SARS-CoV-2 Nucleoprotein Modulating its Phosphoregulation.
J.Mol.Biol., 435, 2023
|
|
7QIP
 
 | | SARS-CoV-2 Nucleocapsid phosphopeptide 201-210 bound to human 14-3-3 sigma | | Descriptor: | 14-3-3 protein sigma, ARG-GLY-TPO-SER-PRO-ALA-ARG-MET, CHLORIDE ION | | Authors: | Sluchanko, N.N, Tugaeva, K.V, Smith, J.L.R, Antson, A.A. | | Deposit date: | 2021-12-15 | | Release date: | 2021-12-29 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Human 14-3-3 Proteins Site-selectively Bind the Mutational Hotspot Region of SARS-CoV-2 Nucleoprotein Modulating its Phosphoregulation.
J.Mol.Biol., 435, 2023
|
|
4XP7
 
 | | Crystal structure of Human tRNA dihydrouridine synthase 2 | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, tRNA-dihydrouridine(20) synthase [NAD(P)+]-like | | Authors: | Whelan, F, Jenkins, H.T, Griffiths, S, Byrne, R.T, Dodson, E.J, Antson, A.A. | | Deposit date: | 2015-01-16 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From bacterial to human dihydrouridine synthase: automated structure determination.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
