1CEM
 
 | | ENDOGLUCANASE A (CELA) CATALYTIC CORE, RESIDUES 33-395 | | Descriptor: | CELLULASE CELA (1,4-BETA-D-GLUCAN-GLUCANOHYDROLASE) | | Authors: | Alzari, P.M. | | Deposit date: | 1995-12-04 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of endoglucanase CelA, a family 8 glycosyl hydrolase from Clostridium thermocellum.
Structure, 4, 1996
|
|
1XYZ
 
 | |
1CLC
 
 | |
1GHL
 
 | |
1HHL
 
 | |
1AOH
 
 | |
1CEC
 
 | |
6CAC
 
 | | Crystal structure of NDM-1 metallo-beta-lactamase harboring an insertion of a Pro residue in L3 loop | | Descriptor: | CADMIUM ION, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Alzari, P.M, Giannini, E, Palacios, A, Mojica, M, Bonomo, R, Llarrull, L, Vila, A. | | Deposit date: | 2018-01-30 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Reaction Mechanism of Metallo-beta-Lactamases Is Tuned by the Conformation of an Active-Site Mobile Loop.
Antimicrob. Agents Chemother., 63, 2019
|
|
1F4Y
 
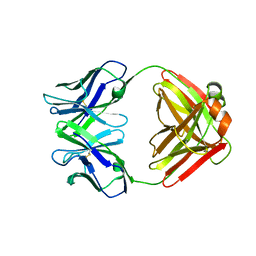 | | CRYSTAL STRUCTURE OF AN ANTI-CARBOHYDRATE ANTIBODY DIRECTED AGAINST VIBRIO CHOLERAE O1 IN COMPLEX WITH ANTIGEN | | Descriptor: | 4,6-dideoxy-4-{[(2R)-2,4-dihydroxybutanoyl]amino}-2-O-methyl-alpha-D-mannopyranose-(1-2)-methyl 4,6-dideoxy-4-{[(2R)-2,4-dihydroxybutanoyl]amino}-alpha-D-mannopyranoside, ANTIBODY S-20-4, FAB FRAGMENT, ... | | Authors: | Alzari, P.M, Souchon, H. | | Deposit date: | 2000-06-10 | | Release date: | 2000-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of an anti-carbohydrate antibody directed against Vibrio cholerae O1 in complex with antigen: molecular basis for serotype specificity.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1F4W
 
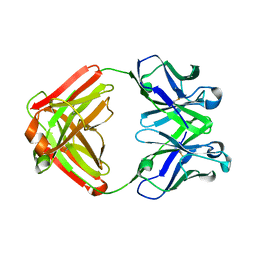 | |
1F4X
 
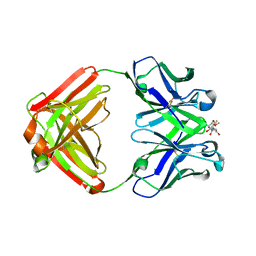 | |
7A1D
 
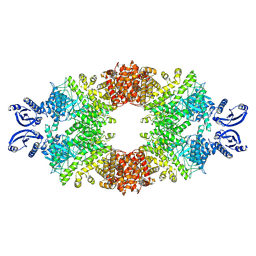 | | Cryo-EM map of the large glutamate dehydrogenase composed of 180 kDa subunits from Mycobacterium smegmatis (open conformation) | | Descriptor: | NAD-specific glutamate dehydrogenase | | Authors: | Lazaro, M, Melero, R, Huet, C, Lopez-Alonso, J.P, Delgado, S, Dodu, A, Bruch, E.M, Abriata, L.A, Alzari, P.M, Valle, M, Lisa, M.N. | | Deposit date: | 2020-08-12 | | Release date: | 2021-06-09 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | 3D architecture and structural flexibility revealed in the subfamily of large glutamate dehydrogenases by a mycobacterial enzyme.
Commun Biol, 4, 2021
|
|
6Z0F
 
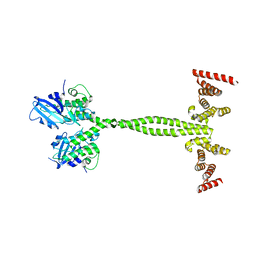 | | Crystal structure of the membrane pseudokinase YukC/EssB from Bacillus subtilis T7SS | | Descriptor: | ESX secretion system protein YukC | | Authors: | Tassinari, M, Bellinzoni, M, Alzari, P.M, Fronzes, R, Gubellini, F. | | Deposit date: | 2020-05-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | The Antibacterial Type VII Secretion System of Bacillus subtilis: Structure and Interactions of the Pseudokinase YukC/EssB.
Mbio, 13, 2022
|
|
8BVE
 
 | | MoeA2 from Corynebacterium glutamicum | | Descriptor: | CITRIC ACID, Molybdopterin molybdenumtransferase, SODIUM ION | | Authors: | Martinez, M, Haouz, A, Wehenkel, A.M, Alzari, P.M. | | Deposit date: | 2022-12-03 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Eukaryotic-like gephyrin and cognate membrane receptor coordinate corynebacterial cell division and polar elongation.
Nat Microbiol, 8, 2023
|
|
8BVF
 
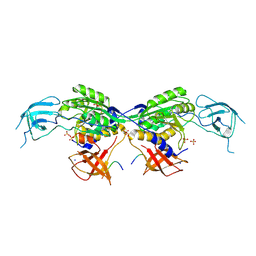 | | MoeA2 from Corynebacterium glutamicum in complex with FtsZ-CTD | | Descriptor: | Cell division protein FtsZ, Molybdopterin molybdenumtransferase, SODIUM ION, ... | | Authors: | Martinez, M, Haouz, A, Wehenkel, A.M, Alzari, P.M. | | Deposit date: | 2022-12-03 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Eukaryotic-like gephyrin and cognate membrane receptor coordinate corynebacterial cell division and polar elongation.
Nat Microbiol, 8, 2023
|
|
3GIF
 
 | | Crystal structure of DesKC_H188E in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Sensor histidine kinase desK | | Authors: | Trajtenberg, F, Albanesi, D, Alzari, P.M, Buschiazzo, A, de Mendoza, D. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural plasticity and catalysis regulation of a thermosensor histidine kinase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GIE
 
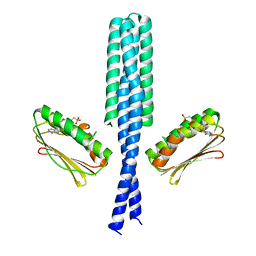 | | Crystal structure of DesKC_H188E in complex with AMP-PCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Sensor histidine kinase desK | | Authors: | Trajtenberg, F, Albanesi, D, Alzari, P.M, Buschiazzo, A, de Mendoza, D. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural plasticity and catalysis regulation of a thermosensor histidine kinase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1MS1
 
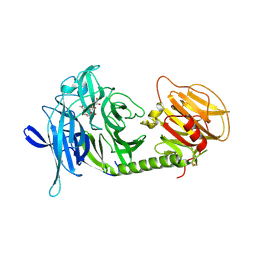 | | Monoclinic form of Trypanosoma cruzi trans-sialidase, in complex with 3-deoxy-2,3-dehydro-N-acetylneuraminic acid (DANA) | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1MZ5
 
 | | Trypanosoma rangeli sialidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, sialidase | | Authors: | Buschiazzo, A, Tavares, G.A, Campetella, O, Spinelli, S, Cremona, M.L, Paris, G, Amaya, M.F, Frasch, A.C.C, Alzari, P.M. | | Deposit date: | 2002-10-05 | | Release date: | 2002-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of sialyltransferase activity in trypanosomal sialidases
Embo J., 19, 2000
|
|
9FM5
 
 | | PvSub1 Catalytic Domain in Complex with Peptidomimetic Inhibitor (AL-97) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5,6-DIHYDRO-BENZO[H]CINNOLIN-3-YLAMINE, CALCIUM ION, ... | | Authors: | Batista, F.A, Martinez, M, Bouillon, A, Mechaly, A, Alzari, P.M, Haouz, A, Barale, J.C. | | Deposit date: | 2024-06-05 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Towards Improved Peptidic alpha-Ketoamide Inhibitors of the Plasmodial Subtilisin-Like SUB1: Exploration of N-Terminal Extensions and Cyclic Constraints.
Chemmedchem, 20, 2025
|
|
8AUC
 
 | | Structure of peptidoglycan hydrolase Cg1735 from Corynebacterium glutamicum, trigonal crystal form | | Descriptor: | Cell wall-associated hydrolases (Invasion-associated proteins), TETRACHLOROPLATINATE(II) | | Authors: | Gaday, Q, Wehenkel, A.M, Alzari, P.M, Legrand, P. | | Deposit date: | 2022-08-25 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | FtsEX-independent control of RipA-mediated cell separation in Corynebacteriales.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8AU6
 
 | |
8AUD
 
 | |
8P5R
 
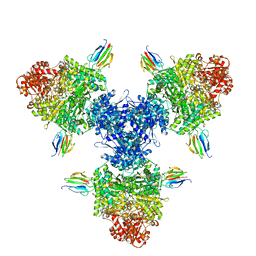 | | Crystal structure of full-length, homohexameric 2-oxoglutarate dehydrogenase KGD from Mycobacterium smegmatis in complex with GarA | | Descriptor: | CALCIUM ION, Glycogen accumulation regulator GarA, MAGNESIUM ION, ... | | Authors: | Wagner, T, Mechaly, A.M, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.562 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
7JSR
 
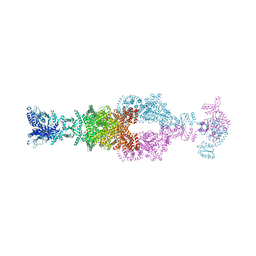 | | Crystal structure of the large glutamate dehydrogenase composed of 180 kDa subunits from Mycobacterium smegmatis | | Descriptor: | NAD-specific glutamate dehydrogenase | | Authors: | Lazaro, M, Melero, R, Huet, C, Lopez-Alonso, J.P, Delgado, S, Dodu, A, Bruch, E.M, Abriata, L.A, Alzari, P.M, Valle, M, Lisa, M.N. | | Deposit date: | 2020-08-15 | | Release date: | 2021-06-09 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (6.27 Å) | | Cite: | 3D architecture and structural flexibility revealed in the subfamily of large glutamate dehydrogenases by a mycobacterial enzyme.
Commun Biol, 4, 2021
|
|
