7EKH
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with human ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | 著者 | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2021-04-05 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKG
 
 | | Structure of SARS-CoV-2 Beta variant spike receptor-binding domain complexed with human ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2021-04-05 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKC
 
 | | Structure of SARS-CoV-2 Gamma variant spike receptor-binding domain complexed with human ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2021-04-05 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
1QK6
 
 | | Solution structure of huwentoxin-I by NMR | | 分子名称: | HUWENTOXIN-I | | 著者 | Qu, Y, Liang, S, Ding, J, Liu, X, Zhang, R, Gu, X. | | 登録日 | 1999-07-10 | | 公開日 | 1999-08-20 | | 最終更新日 | 2019-01-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Proton Nuclear Magnetic Resonance Studies on Huwentoxin-I from the Venom of the Spider Selenocosmia Huwena:2.Three-Dimensional Structure in Solution
J.Protein Chem., 16, 1997
|
|
7DDZ
 
 | | The Crystal Structure of Human Neuropeptide Y Y2 Receptor with JNJ-31020028 | | 分子名称: | FLAVIN MONONUCLEOTIDE, Human Neuropeptide Y Y2 Receptor fusion protein, ~{N}-[4-[4-[(1~{S})-2-(diethylamino)-2-oxidanylidene-1-phenyl-ethyl]piperazin-1-yl]-3-fluoranyl-phenyl]-2-pyridin-3-yl-benzamide | | 著者 | Tang, T, Han, S, Zhao, Q, Wu, B. | | 登録日 | 2020-10-30 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for ligand recognition of the neuropeptide Y Y 2 receptor.
Nat Commun, 12, 2021
|
|
4OU4
 
 | | Crystal structure of esterase rPPE mutant S159A complexed with (S)-Ac-CPA | | 分子名称: | (2S)-(acetyloxy)(2-chlorophenyl)ethanoic acid, Alpha/beta hydrolase fold-3 domain protein | | 著者 | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | 登録日 | 2014-02-15 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
7VFX
 
 | | The structure of Formyl Peptide Receptor 1 in complex with Gi and peptide agonist fMIFL | | 分子名称: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wang, X.K, Chen, G, Liao, Q.W, Du, Y, Hu, H.L, Ye, D.Q. | | 登録日 | 2021-09-14 | | 公開日 | 2022-09-21 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis for recognition of N-formyl peptides as pathogen-associated molecular patterns.
Nat Commun, 13, 2022
|
|
4OB8
 
 | | Crystal structure of a novel thermostable esterase from Pseudomonas putida ECU1011 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | 著者 | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | 登録日 | 2014-01-07 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4OB7
 
 | | Crystal structure of esterase rPPE mutant W187H | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | 著者 | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | 登録日 | 2014-01-07 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4OU5
 
 | | Crystal structure of esterase rPPE mutant S159A/W187H | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | 著者 | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | 登録日 | 2014-02-15 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4RNC
 
 | | Crystal structure of an esterase RhEst1 from Rhodococcus sp. ECU1013 | | 分子名称: | Esterase, PHOSPHATE ION | | 著者 | Dou, S, Kong, X.D, Xu, J.H, Zhou, J. | | 登録日 | 2014-10-23 | | 公開日 | 2015-10-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Substrate channel evolution of an esterase for the synthesis of Cilastatin
CATALYSIS SCIENCE AND TECHNOLOGY, 5, 2015
|
|
7DDP
 
 | | Cryo-EM structure of human ACE2 and GX/P2V/2017 RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | 著者 | Niu, S, Wang, J, Wang, H.W, Qi, J.X, Wang, Q.H, Gao, G.F. | | 登録日 | 2020-10-29 | | 公開日 | 2021-05-19 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin.
Embo J., 40, 2021
|
|
7DDO
 
 | | Cryo-EM structure of human ACE2 and GD/1/2019 RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | 著者 | Niu, S, Wang, J, Wang, H.W, Qi, J.X, Wang, Q.H, Gao, G.F. | | 登録日 | 2020-10-29 | | 公開日 | 2021-05-19 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin.
Embo J., 40, 2021
|
|
5X9C
 
 | |
7DHX
 
 | | Crystal structure of SARS-CoV-2 RBD binding to pangolin ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ZINC ION, ... | | 著者 | Wang, Q.H, Qi, J.X, Wu, L.L. | | 登録日 | 2020-11-17 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Molecular basis of pangolin ACE2 engaged by COVID-19 virus
Chin.Sci.Bull., 66, 2021
|
|
5X9B
 
 | |
7E3L
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 58G6 heavy chain, 58G6 light chain, ... | | 著者 | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2021-02-09 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
7E3K
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | 分子名称: | 13G9 heavy chain, 13G9 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2021-02-09 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
8HYA
 
 | |
8HNM
 
 | | CXCR3-DNGi complex activated by VUF11222 | | 分子名称: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Jiao, H.Z, Hu, H.L. | | 登録日 | 2022-12-08 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HNN
 
 | | Structure of CXCR3 complexed with antagonist SCH546738 | | 分子名称: | 3-azanyl-6-chloranyl-5-[(3S)-4-[1-[(4-chlorophenyl)methyl]piperidin-4-yl]-3-ethyl-piperazin-1-yl]pyrazine-2-carboxamide, CHOLESTEROL, Nb6, ... | | 著者 | Jiao, H.Z, Hu, H.L. | | 登録日 | 2022-12-08 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HNK
 
 | | CXCR3-DNGi complex activated by CXCL11 | | 分子名称: | C-X-C motif chemokine 11, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Jiao, H.Z, Hu, H.L. | | 登録日 | 2022-12-08 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HNL
 
 | | CXCR3-DNGi complex activated by PS372424 | | 分子名称: | (3S)-N-[(2S)-5-carbamimidamido-1-(cyclohexylmethylamino)-1-oxidanylidene-pentan-2-yl]-2-(4-oxidanylidene-4-phenyl-butanoyl)-3,4-dihydro-1H-isoquinoline-3-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Jiao, H.Z, Hu, H.L. | | 登録日 | 2022-12-08 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HW3
 
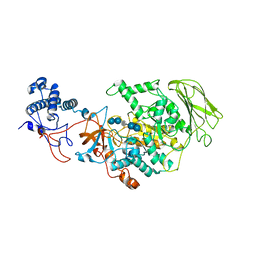 | | Limosilactobacillus reuteri N1 GtfB-acarbose | | 分子名称: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, GLYCEROL, SODIUM ION, ... | | 著者 | Dong, J.J, Bai, Y.X. | | 登録日 | 2022-12-28 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Insights into the Structure-Function Relationship of GH70 GtfB alpha-Glucanotransferases from the Crystal Structure and Molecular Dynamic Simulation of a Newly Characterized Limosilactobacillus reuteri N1 GtfB Enzyme.
J.Agric.Food Chem., 72, 2024
|
|
8HWK
 
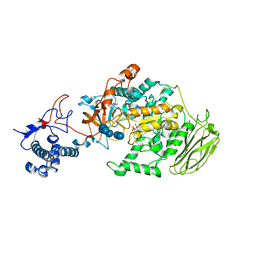 | | Limosilactobacillus reuteri N1 GtfB-maltohexaose | | 分子名称: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | 著者 | Dong, J.J, Bai, Y.X. | | 登録日 | 2022-12-30 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Insights into the Structure-Function Relationship of GH70 GtfB alpha-Glucanotransferases from the Crystal Structure and Molecular Dynamic Simulation of a Newly Characterized Limosilactobacillus reuteri N1 GtfB Enzyme.
J.Agric.Food Chem., 72, 2024
|
|
