1A66
 
 | | SOLUTION NMR STRUCTURE OF THE CORE NFATC1/DNA COMPLEX, 18 STRUCTURES | | 分子名称: | CORE NFATC1, DNA (5'-D(*CP*AP*AP*TP*TP*TP*TP*CP*CP*TP*CP*G)-3'), DNA (5'-D(*CP*GP*AP*GP*GP*AP*AP*AP*AP*TP*TP*G)-3') | | 著者 | Zhou, P, Sun, L.J, Doetsch, V, Wagner, G, Verdine, G.L. | | 登録日 | 1998-03-06 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the core NFATC1/DNA complex.
Cell(Cambridge,Mass.), 92, 1998
|
|
1IBX
 
 | | NMR STRUCTURE OF DFF40 AND DFF45 N-TERMINAL DOMAIN COMPLEX | | 分子名称: | CHIMERA OF IGG BINDING PROTEIN G AND DNA FRAGMENTATION FACTOR 45, DNA FRAGMENTATION FACTOR 40 | | 著者 | Zhou, P, Lugovskoy, A.A, McCarty, J.S, Li, P, Wagner, G. | | 登録日 | 2001-03-29 | | 公開日 | 2001-05-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of DFF40 and DFF45 N-terminal domain complex and mutual chaperone activity of DFF40 and DFF45.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1C15
 
 | | SOLUTION STRUCTURE OF APAF-1 CARD | | 分子名称: | APOPTOTIC PROTEASE ACTIVATING FACTOR 1 | | 著者 | Zhou, P, Chou, J, Olea, R.S, Yuan, J, Wagner, G. | | 登録日 | 1999-07-20 | | 公開日 | 1999-09-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of Apaf-1 CARD and its interaction with caspase-9 CARD: a structural basis for specific adaptor/caspase interaction.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3VM7
 
 | | Structure of an Alpha-Amylase from Malbranchea cinnamomea | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | 著者 | Zhou, P, Hu, S.Q, Zhou, Y, Han, P, Yang, S.Q, Jiang, Z.Q. | | 登録日 | 2011-12-09 | | 公開日 | 2013-05-29 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | A Novel Multifunctional alpha-Amylase from the Thermophilic Fungus Malbranchea cinnamomea: Biochemical Characterization and Three-Dimensional Structure.
Appl Biochem Biotechnol., 170, 2013
|
|
4K3A
 
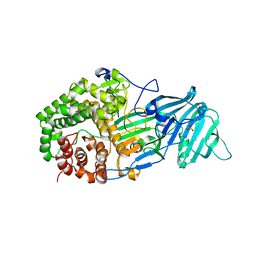 | | The structure of a glycoside hydrolase family 81 endo-[beta]-1,3-glucanase | | 分子名称: | SULFATE ION, glycoside hydrolase family 81 endo-beta-1,3-glucanase | | 著者 | Jiang, Z.Q, Zhou, P, Chen, Z.Z, Yan, Q.J, Yang, S.Q, Hilgenfeld, R. | | 登録日 | 2013-04-10 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The structure of a glycoside hydrolase family 81
endo-[beta]-1,3-glucanase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4K35
 
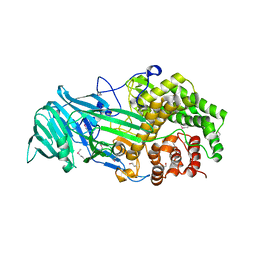 | | The structure of a glycoside hydrolase family 81 endo-[beta]-1,3-glucanase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, glycoside hydrolase family 81 endo-beta-1,3-glucanase | | 著者 | Jiang, Z.Q, Zhou, P, Chen, Z.Z, Yan, Q.J, Yang, S.Q, Hilgenfeld, R. | | 登録日 | 2013-04-10 | | 公開日 | 2013-10-02 | | 実験手法 | X-RAY DIFFRACTION (2.003 Å) | | 主引用文献 | The structure of a glycoside hydrolase family 81
endo-[beta]-1,3-glucanase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2I5O
 
 | |
4LYR
 
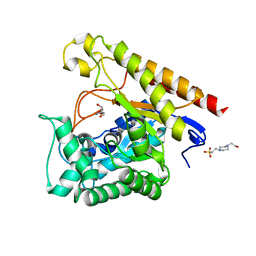 | | Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E301A mutant | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Exo-beta-1,4-mannosidase | | 著者 | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | 登録日 | 2013-07-31 | | 公開日 | 2014-08-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LYP
 
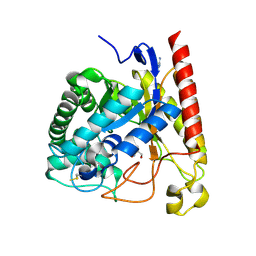 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Exo-beta-1,4-mannosidase, GUANIDINE | | 著者 | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | 登録日 | 2013-07-31 | | 公開日 | 2014-08-06 | | 最終更新日 | 2014-11-26 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LYQ
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E202A mutant | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Exo-beta-1,4-mannosidase, ... | | 著者 | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | 登録日 | 2013-07-31 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NRS
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannobiose | | 分子名称: | Exo-beta-1,4-mannosidase, beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | 著者 | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | 登録日 | 2013-11-27 | | 公開日 | 2014-11-19 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NRR
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannosyl-fructose | | 分子名称: | Exo-beta-1,4-mannosidase, beta-D-mannopyranose-(1-4)-beta-D-fructofuranose | | 著者 | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | 登録日 | 2013-11-27 | | 公開日 | 2014-11-19 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2A7O
 
 | | Solution Structure of the hSet2/HYPB SRI domain | | 分子名称: | Huntingtin interacting protein B | | 著者 | Li, M, Phatnani, H.P, Guan, Z, Sage, H, Greenleaf, A, Zhou, P. | | 登録日 | 2005-07-05 | | 公開日 | 2005-11-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the Set2 Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6PIB
 
 | | Structure of the Klebsiella pneumoniae LpxH-AZ1 complex | | 分子名称: | 1-[5-({4-[3-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)-2,3-dihydro-1H-indol-1-yl]ethan-1-one, MANGANESE (II) ION, TETRAETHYLENE GLYCOL, ... | | 著者 | Cho, J, Zhou, P. | | 登録日 | 2019-06-26 | | 公開日 | 2020-02-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4LCH
 
 | | Crystal structure of the Pseudomonas aeruginosa LPXC/LPC-051 complex | | 分子名称: | (betaS)-Nalpha-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N,beta-dihydroxy-beta-methyl-L-tyrosinamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2013-06-21 | | 公開日 | 2013-08-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.596 Å) | | 主引用文献 | Synthesis, Structure, and Antibiotic Activity of Aryl-Substituted LpxC Inhibitors.
J.Med.Chem., 56, 2013
|
|
4LCF
 
 | | Crystal structure of the Pseudomonas aeruginosa LPXC/LPC-014 complex | | 分子名称: | NITRATE ION, Nalpha-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N-hydroxy-L-histidinamide, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2013-06-21 | | 公開日 | 2013-08-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | Synthesis, Structure, and Antibiotic Activity of Aryl-Substituted LpxC Inhibitors.
J.Med.Chem., 56, 2013
|
|
4LKV
 
 | | Determinants of lipid substrate and membrane binding for the tetraacyldisaccharide-1-phosphate 4 -kinase LpxK | | 分子名称: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-deoxy-3-O-[(3R)-3-hydroxytetradecanoyl]-2-{[(3R)-3-hydroxytetradecanoyl]amino}-4-O-phosphono-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Emptage, R.P, Tonthat, N.K, York, J.D, Schumacher, M.A, Zhou, P. | | 登録日 | 2013-07-08 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.5109 Å) | | 主引用文献 | Structural Basis of Lipid Binding for the Membrane-embedded Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK.
J.Biol.Chem., 289, 2014
|
|
2KIQ
 
 | | Solution structure of the FF Domain 2 of human transcription elongation factor CA150 | | 分子名称: | Transcription elongation regulator 1 | | 著者 | Zeng, J, Boyles, J, Tripathy, C, Yan, A, Zhou, P, Donald, B.R. | | 登録日 | 2009-05-07 | | 公開日 | 2009-07-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | High-resolution protein structure determination starting with a global fold calculated from exact solutions to the RDC equations.
J.Biomol.Nmr, 45, 2009
|
|
5XBZ
 
 | | Crystal structure of GH family 81 beta-1,3-glucanase from Rhizomucr miehei complexed with laminaripentaose | | 分子名称: | Endo-beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | 著者 | Yang, S, Qin, Z, Zhou, P, Yan, Q, Jiang, Z. | | 登録日 | 2017-03-21 | | 公開日 | 2018-03-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Catalytic mechanism of glycoside hydrolase family 81 beta-1,3-glucanase
To Be Published
|
|
3WNV
 
 | | Crystal structure of a glyoxylate reductase from Paecilomyes thermophila | | 分子名称: | SULFATE ION, glyoxylate reductase | | 著者 | Duan, X, Hu, S, Zhou, P, Zhou, Y, Jiang, Z. | | 登録日 | 2013-12-17 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Characterization and crystal structure of a first fungal glyoxylate reductase from Paecilomyes thermophila
Enzyme.Microb.Technol., 60, 2014
|
|
1D4B
 
 | | CIDE-N DOMAIN OF HUMAN CIDE-B | | 分子名称: | HUMAN CELL DEATH-INDUCING EFFECTOR B | | 著者 | Lugovskoy, A, Zhou, P, Chou, J, McCarty, J, Li, P, Wagner, G. | | 登録日 | 1999-10-02 | | 公開日 | 1999-12-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the CIDE-N domain of CIDE-B and a model for CIDE-N/CIDE-N interactions in the DNA fragmentation pathway of apoptosis.
Cell(Cambridge,Mass.), 99, 1999
|
|
2N1T
 
 | | Dynamic binding mode of a synaptotagmin-1-SNARE complex in solution | | 分子名称: | Synaptosomal-associated protein 25, Synaptotagmin-1, Syntaxin-1A, ... | | 著者 | Brewer, K, Bacaj, T, Cavalli, A, Camilloni, C, Swarbrick, J, Liu, J, Zhou, A, Zhou, P, Barlow, N, Xu, J, Seven, A, Prinslow, E, Voleti, R, Haussinger, D, Bonvin, A, Tomchick, D, Vendruscolo, M, Graham, B, Sudhof, T, Rizo, J. | | 登録日 | 2015-04-21 | | 公開日 | 2015-06-03 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Dynamic binding mode of a Synaptotagmin-1-SNARE complex in solution.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1U8B
 
 | | Crystal structure of the methylated N-ADA/DNA complex | | 分子名称: | 5'-D(*AP*AP*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*T)-3', 5'-D(*TP*AP*AP*AP*TP*T)-3', 5'-D(P*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*AP*T)-3', ... | | 著者 | He, C, Hus, J.-C, Sun, L.J, Zhou, P, Norman, D.P.G, Dotsch, V, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | 登録日 | 2004-08-05 | | 公開日 | 2005-10-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A methylation-dependent electrostatic switch controls DNA repair and transcriptional activation by E. coli ada.
Mol.Cell, 20, 2005
|
|
1NFA
 
 | | HUMAN TRANSCRIPTION FACTOR NFATC DNA BINDING DOMAIN, NMR, 10 STRUCTURES | | 分子名称: | HUMAN TRANSCRIPTION FACTOR NFATC1 | | 著者 | Wolfe, S.A, Zhou, P, Dotsch, V, Chen, L, You, A, Ho, S.N, Crabtree, G.R, Wagner, G, Verdine, G.L. | | 登録日 | 1997-01-18 | | 公開日 | 1997-04-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Unusual Rel-like architecture in the DNA-binding domain of the transcription factor NFATc.
Nature, 385, 1997
|
|
2MBB
 
 | |
