7F5W
 
 | |
7FCQ
 
 | |
7FCP
 
 | |
2MDC
 
 | | Solution structure of the WW domain of HYPB | | 分子名称: | Histone-lysine N-methyltransferase SETD2 | | 著者 | Gao, Y.G. | | 登録日 | 2013-09-10 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Autoinhibitory structure of the WW domain of HYPB/SETD2 regulates its interaction with the proline-rich region of huntingtin
Structure, 22, 2014
|
|
7FBJ
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing nanobody 17F6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, ... | | 著者 | Zhu, J, Xu, T, Feng, B, Liu, J. | | 登録日 | 2021-07-11 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
7FBK
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain N501Y mutant in complex with neutralizing nanobody 20G6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, Spike protein S1 | | 著者 | Zhu, J, Xu, T, Feng, B, Liu, J. | | 登録日 | 2021-07-11 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
6KXW
 
 | | Crystal structure of human aquaporin AQP7 in bound to glycerol | | 分子名称: | Aquaporin-7, GLYCEROL | | 著者 | Zhang, L, Yao, D, Zhou, F, Zhang, Q, Zhou, L, Cao, Y. | | 登録日 | 2019-09-13 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | The structural basis for glycerol permeation by human AQP7
Sci Bull (Beijing), 66, 2020
|
|
6LYC
 
 | | Crystal structure of the NOD SIRPa complex with D4-2 | | 分子名称: | ACETIC ACID, D4-2, SIRPa of the NOD mouse strain | | 著者 | Murata, Y, Matsuda, M, Nakagawa, A, Matozaki, T. | | 登録日 | 2020-02-14 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Macrocyclic Peptide-Mediated Blockade of the CD47-SIRP alpha Interaction as a Potential Cancer Immunotherapy.
Cell Chem Biol, 27, 2020
|
|
4LCG
 
 | | Crystal structure of the Pseudomonas aeruginosa LPXC/LPC-050 complex | | 分子名称: | (betaS)-Nalpha-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N,beta-dihydroxy-L-tyrosinamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2013-06-21 | | 公開日 | 2013-08-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.568 Å) | | 主引用文献 | Synthesis, Structure, and Antibiotic Activity of Aryl-Substituted LpxC Inhibitors.
J.Med.Chem., 56, 2013
|
|
7DW0
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp14|15 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp14/15 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW6
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp15|16 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp15/16 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVP
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp4|5 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp4/5 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-14 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVW
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp5|6 peptidyl substrate | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, nsp5/6 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVY
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp9|10 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp9/10 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVX
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp6|7 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp6/7 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7E5X
 
 | |
7WJY
 
 | | Omicron spike trimer with 6m6 antibody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | 著者 | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | 登録日 | 2022-01-08 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
7WK0
 
 | | Local refine of Omicron spike bitrimer with 6m6 antibody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | 著者 | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | 登録日 | 2022-01-08 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
7WJZ
 
 | | Omicron Spike bitrimer with 6m6 antibody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | 著者 | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | 登録日 | 2022-01-08 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
4LCH
 
 | | Crystal structure of the Pseudomonas aeruginosa LPXC/LPC-051 complex | | 分子名称: | (betaS)-Nalpha-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N,beta-dihydroxy-beta-methyl-L-tyrosinamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2013-06-21 | | 公開日 | 2013-08-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.596 Å) | | 主引用文献 | Synthesis, Structure, and Antibiotic Activity of Aryl-Substituted LpxC Inhibitors.
J.Med.Chem., 56, 2013
|
|
4LCF
 
 | | Crystal structure of the Pseudomonas aeruginosa LPXC/LPC-014 complex | | 分子名称: | NITRATE ION, Nalpha-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N-hydroxy-L-histidinamide, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2013-06-21 | | 公開日 | 2013-08-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | Synthesis, Structure, and Antibiotic Activity of Aryl-Substituted LpxC Inhibitors.
J.Med.Chem., 56, 2013
|
|
7W9N
 
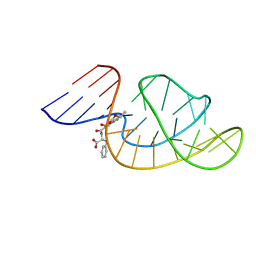 | | THE STRUCTURE OF OBA33-OTA COMPLEX | | 分子名称: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, OTA DNA APTAMER (33-MER) | | 著者 | Xu, G.H, Li, C.G. | | 登録日 | 2021-12-10 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Insights into the Mechanism of High-Affinity Binding of Ochratoxin A by a DNA Aptamer.
J.Am.Chem.Soc., 144, 2022
|
|
7X2H
 
 | |
7XD2
 
 | |
7Y7Q
 
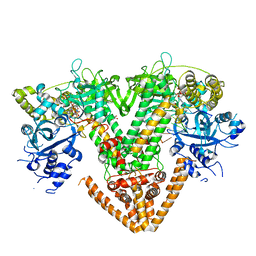 | | QDE-1 in complex with RNA template, RNA primer and 3'-dGTP | | 分子名称: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Cui, R.X, Gan, J.H, Ma, J.B. | | 登録日 | 2022-06-22 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural insights into the dual activities of the two-barrel RNA polymerase QDE-1.
Nucleic Acids Res., 50, 2022
|
|
