2QG1
 
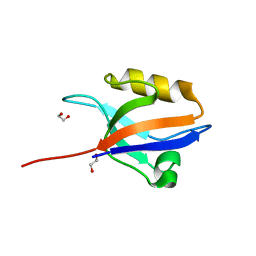 | | Crystal structure of the 11th PDZ domain of MPDZ (MUPP1) | | Descriptor: | 1,2-ETHANEDIOL, Multiple PDZ domain protein | | Authors: | Papagrigoriou, E, Salah, E, Phillips, C, Savitsky, P, Boisguerin, P, Oschkinat, H, Gileadi, C, Yang, X, Elkins, J.M, Ugochukwu, E, Bunkoczi, G, Uppenberg, J, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the 11th PDZ domain of MPDZ (MUPP1).
To be Published
|
|
2QME
 
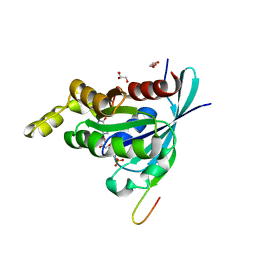 | | Crystal structure of human RAC3 in complex with CRIB domain of human p21-activated kinase 1 (PAK1) | | Descriptor: | CRIB domain of the Serine/threonine-protein kinase PAK 1, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ugochukwu, E, Yang, X, Elkins, J.M, Burgess-Brown, N, Bunkoczi, G, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Knapp, S, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-16 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of the human RAC3 in complex with the CRIB domain of human p21-activated kinase 1 (PAK1).
To be Published
|
|
5JMT
 
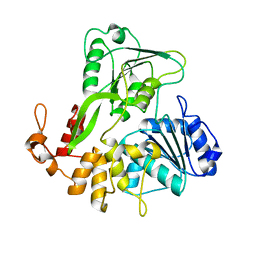 | | Crystal structure of Zika virus NS3 helicase | | Descriptor: | NS3 helicase | | Authors: | Tian, H, Ji, X, Yang, X, Xie, W, Yang, K, Chen, C, Wu, C, Chi, H, Mu, Z, Wang, Z, Yang, H. | | Deposit date: | 2016-04-29 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | The crystal structure of Zika virus helicase: basis for antiviral drug design
Protein Cell, 7, 2016
|
|
7YR7
 
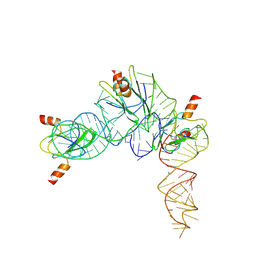 | | Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with three RsmA protein dimers | | Descriptor: | RsmZ RNA (118-MER), Translational regulator CsrA | | Authors: | Jia, X, Pan, Z, Yuan, Y, Luo, B, Luo, Y, Mukherjee, S, Jia, G, Liu, L, Ling, X, Yang, X, Wu, Y, Liu, T, Miao, Z, Wei, X, Bujnicki, J.M, Zhao, K, Su, Z. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of sRNA RsmZ regulation of Pseudomonas aeruginosa virulence.
Cell Res., 33, 2023
|
|
7YR6
 
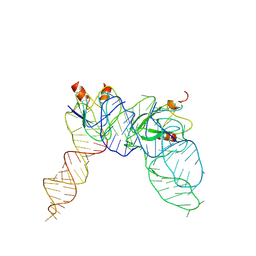 | | Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with two RsmA protein dimers | | Descriptor: | RsmZ RNA, Translational regulator CsrA | | Authors: | Jia, X, Pan, Z, Yuan, Y, Luo, B, Luo, Y, Mukherjee, S, Jia, G, Ling, X, Yang, X, Wu, Y, Liu, T, Wei, X, Bujnick, J.M, Zhao, K, Su, Z. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of sRNA RsmZ regulation of Pseudomonas aeruginosa virulence.
Cell Res., 33, 2023
|
|
3SLU
 
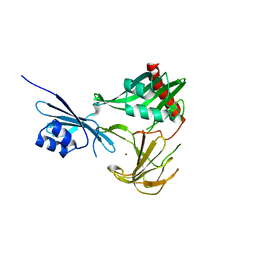 | | Crystal structure of NMB0315 | | Descriptor: | M23 peptidase domain protein, NICKEL (II) ION | | Authors: | Shen, Y, Wang, X, Yang, X, Xu, H. | | Deposit date: | 2011-06-26 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of outer membrane protein NMB0315 from Neisseria meningitidis.
Plos One, 6, 2011
|
|
1NOQ
 
 | | e-motif structure | | Descriptor: | 5'-D(*CP*CP*GP*CP*CP*G)-3' | | Authors: | Zheng, M, Huang, X, Smith, G.K, Yang, X, Gao, X. | | Deposit date: | 2003-01-16 | | Release date: | 2003-02-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Genetically unstable CXG repeats are structurally dynamic and have a high propensity for folding.
An NMR and UV spectroscopic study.
J.Mol.Biol., 264, 1996
|
|
4ZM6
 
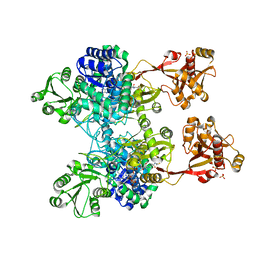 | | A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase | | Descriptor: | ACETYL COENZYME *A, N-acetyl-beta-D glucosaminidase, SULFATE ION | | Authors: | Qin, Z, Xiao, Y, Yang, X, Jiang, Z, Yang, S, Mesters, J.R. | | Deposit date: | 2015-05-02 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain glycoside hydrolase family 3 beta-N-acetylglucosaminidase
Sci Rep, 5, 2015
|
|
2C2H
 
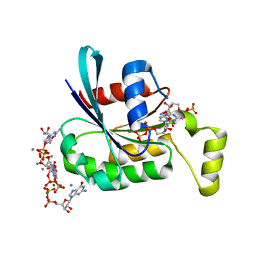 | | CRYSTAL STRUCTURE OF THE HUMAN RAC3 IN COMPLEX WITH GDP | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Debreczeni, J.E, Yang, X, Zao, Y, Elkins, J, Gileadi, C, Burgess, N, Colebrook, S, Gileadi, O, Fedorov, O, Bunkoczi, G, von Delft, F, Doyle, D, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A. | | Deposit date: | 2005-09-29 | | Release date: | 2005-10-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Human Rac3 in Complex with Gdp
To be Published
|
|
2CLS
 
 | | The crystal structure of the human RND1 GTPase in the active GTP bound state | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RHO-RELATED GTP-BINDING PROTEIN RHO6 | | Authors: | Pike, A.C.W, Yang, X, Colebrook, S, Gileadi, O, Sobott, F, Bray, J, Wen Hwa, L, Marsden, B, Zhao, Y, Schoch, G, Elkins, J, Debreczeni, J.E, Turnbull, A.P, von Delft, F, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Doyle, D. | | Deposit date: | 2006-04-28 | | Release date: | 2006-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Crystal Structure of the Human Rnd1 Gtpase in the Active GTP Bound State
To be Published
|
|
2ARH
 
 | | Crystal Structure of a Protein of Unknown Function AQ1966 from Aquifex aeolicus VF5 | | Descriptor: | CALCIUM ION, SELENIUM ATOM, SULFATE ION, ... | | Authors: | Qiu, Y, Kim, Y, Yang, X, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-19 | | Release date: | 2005-10-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal Structure of a Hypothetical Protein Aq_1966 from Aquifex aeolicus VF5
To be Published
|
|
2B05
 
 | | Crystal Structure of 14-3-3 gamma in complex with a phosphoserine peptide | | Descriptor: | 14-3-3 protein gamma, peptide | | Authors: | Papagrigoriou, E, Elkins, J, Arrowsmith, C, Zhao, Y, Debreczeni, E.J, Edwards, A, Weigelt, J, Doyle, D, von Delft, F, Turnbull, A, Yang, X. | | Deposit date: | 2005-09-13 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of 14-3-3 gamma in complex with a phosphoserine peptide
TO BE PUBLISHED
|
|
2ERX
 
 | | Crystal Structure of DiRas2 in Complex With GDP and Inorganic Phosphate | | Descriptor: | GTP-binding protein Di-Ras2, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Papagrigoriou, E, Yang, X, Elkins, J, Niesen, F.E, Burgess, N, Salah, E, Fedorov, O, Ball, L.J, von Delft, F, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Doyle, D. | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of DiRas2
To be Published
|
|
2F5Y
 
 | | Crystal Structure of the PDZ Domain from Human RGS-3 | | Descriptor: | SULFATE ION, regulator of G-protein signalling 3 isoform 1 | | Authors: | Ugochukwu, E, Berridge, G, Johansson, C, Smee, C, Savitsky, P, Burgess, N, Colebrook, S, Yang, X, Elkins, J, Doyle, D, Turnbull, A, Papagrigoriou, E, Debreczeni, J, Bunkoczi, G, Gorrec, F, von Delft, F, Arrowsmith, C, Sundstrom, M, Weigelt, J, Edwards, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-28 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal Structure of the PDZ Domain from Human RGS-3
To be Published
|
|
4Q2J
 
 | | A novel structure-based mechanism for DNA-binding of SATB1 | | Descriptor: | DNA-binding protein SATB1 | | Authors: | Wang, Z, Long, J, Yang, X, Shen, Y. | | Deposit date: | 2014-04-08 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Crystal structure of the ubiquitin-like domain-CUT repeat-like tandem of special AT-rich sequence binding protein 1 (SATB1) reveals a coordinating DNA-binding mechanism.
J.Biol.Chem., 289, 2014
|
|
5EB3
 
 | | VB6-bound protein | | Descriptor: | 4,5-bis(hydroxymethyl)-2-methyl-pyridin-3-ol, SULFATE ION, YfiR | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5EB0
 
 | | crystal form II of YfiB belonging to P41 | | Descriptor: | SULFATE ION, YfiB | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5EB2
 
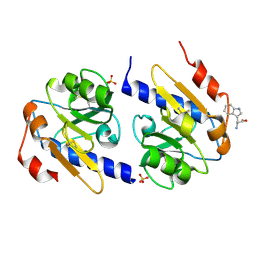 | | Trp-bound YfiR | | Descriptor: | SULFATE ION, TRYPTOPHAN, YfiR | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.709 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5EB1
 
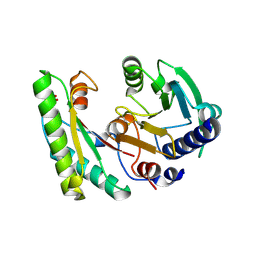 | | the YfiB-YfiR complex | | Descriptor: | SULFATE ION, YfiB, YfiR | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5EAZ
 
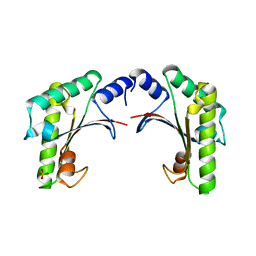 | | crystal form I of YfiB belonging to space groups P21 | | Descriptor: | SULFATE ION, YfiB | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
1K8L
 
 | | XBY6: An analog of CK14 containing 6 dithiophosphate groups | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1K8J
 
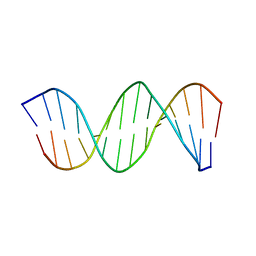 | | NMR STRUCTURE OF THE CK14 DNA DUPLEX: A PORTION OF THE KNOWN NF-kB SEQUENCE CK1 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1K8N
 
 | | NMR structure of the XBY2 DNA duplex, an analog of CK14 containing phosphorodithioate groups at C22 and C24 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
8DQL
 
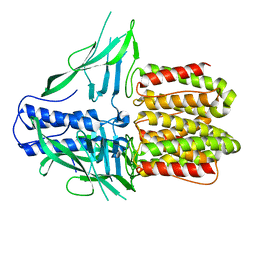 | | CryoEM structure of IglD | | Descriptor: | Secretion system protein | | Authors: | Liu, X, Clemens, D, Lee, B, Yang, X, Zhou, H, Horwitz, M. | | Deposit date: | 2022-07-19 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Atomic Structure of IglD Demonstrates Its Role as a Component of the Baseplate Complex of the Francisella Type VI Secretion System.
Mbio, 13, 2022
|
|
5T4D
 
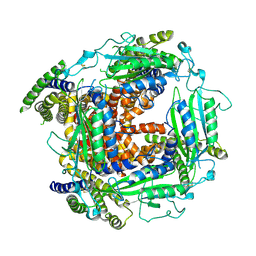 | | Cryo-EM structure of Polycystic Kidney Disease protein 2 (PKD2), residues 198-703 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, hPKD:198-703, Polycystin-2 | | Authors: | Shen, P.S, Yang, X, DeCaen, P.G, Liu, X, Bulkley, D, Clapham, D.E, Cao, E. | | Deposit date: | 2016-08-29 | | Release date: | 2016-11-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Structure of the Polycystic Kidney Disease Channel PKD2 in Lipid Nanodiscs.
Cell, 167, 2016
|
|
