7JQ5
 
 | |
7JQ3
 
 | |
7M2P
 
 | |
7RSM
 
 | |
7BUY
 
 | | The crystal structure of COVID-19 main protease in complex with carmofur | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, hexylcarbamic acid | | 著者 | Zhao, Y, Zhang, B, Jin, Z, Liu, X, Yang, H, Rao, Z. | | 登録日 | 2020-04-08 | | 公開日 | 2020-04-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7BQY
 
 | | THE CRYSTAL STRUCTURE OF COVID-19 MAIN PROTEASE IN COMPLEX WITH AN INHIBITOR N3 at 1.7 angstrom | | 分子名称: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | 著者 | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | 登録日 | 2020-03-26 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
6LU7
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor N3 | | 分子名称: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | 著者 | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | 登録日 | 2020-01-26 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
8WY3
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor 21 | | 分子名称: | Bromodomain-containing protein 4, ~{N}-cyclopropyl-2-[[5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-2-oxidanylidene-pyridin-4-yl]amino]ethanamide | | 著者 | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | 登録日 | 2023-10-30 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
8WXY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor 23 | | 分子名称: | 5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-4-[(2-morpholin-4-yl-2-oxidanylidene-ethyl)amino]pyridin-2-one, Bromodomain-containing protein 4, GLYCEROL | | 著者 | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | 登録日 | 2023-10-30 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
8WY7
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor 22 | | 分子名称: | 2-[[5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-2-oxidanylidene-pyridin-4-yl]amino]-~{N}-(4-oxidanylcyclohexyl)ethanamide, Bromodomain-containing protein 4 | | 著者 | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | 登録日 | 2023-10-30 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
8WYG
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor 22 | | 分子名称: | 2-[[5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-2-oxidanylidene-pyridin-4-yl]amino]-~{N}-(4-oxidanylcyclohexyl)ethanamide, Bromodomain-containing protein 2 | | 著者 | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | 登録日 | 2023-10-30 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.13 Å) | | 主引用文献 | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
5GIX
 
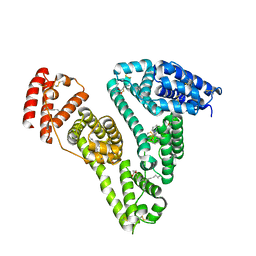 | | Human serum albumin-Palmitic acid-Fe(Hn3piT)Cl2 | | 分子名称: | 14-piperidin-1-yl-11-oxa-13$l^{3}-thia-15,16$l^{4}-diaza-12$l^{3}-ferratetracyclo[8.7.0.0^{2,7}.0^{12,16}]heptadeca-1(10),2(7),3,5,8,13,16-heptaene, PALMITIC ACID, Serum albumin | | 著者 | Yang, F, Qi, J, Wang, T. | | 登録日 | 2016-06-25 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.801 Å) | | 主引用文献 | Developing Anticancer Ferric Prodrugs Based on the N-Donor Residues of Human Serum Albumin Carrier IIA Subdomain
J. Med. Chem., 59, 2016
|
|
4MRO
 
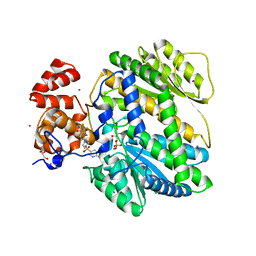 | | Human GKRP bound to AMG-5980 and S6P | | 分子名称: | 2-(4-{4-[(6-aminopyridin-3-yl)sulfonyl]piperazin-1-yl}phenyl)-1,1,1,3,3,3-hexafluoropropan-2-ol, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | 著者 | St Jean, D.J, Ashton, K.S, Bartberger, M.D, Chen, J, Chmait, S, Cupples, R, Galbreath, E, Helmering, J, Jordan, S.R, Liu, L, Kunz, K, Michelsen, K, Nishimura, N, Pennington, L.D, Poon, S.F, Sivits, G, Stec, M.M, Tamayo, N, Van, G, Yang, K, Norman, M.H, Fotsch, C, LLoyd, D.J, Hale, C. | | 登録日 | 2013-09-17 | | 公開日 | 2014-05-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Small molecule disruptors of the glucokinase-glucokinase regulatory protein interaction: 2. Leveraging structure-based drug design to identify analogues with improved pharmacokinetic profiles.
J.Med.Chem., 57, 2014
|
|
2H2Z
 
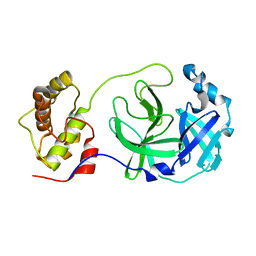 | | Crystal structure of SARS-CoV main protease with authentic N and C-termini | | 分子名称: | Replicase polyprotein 1ab | | 著者 | Yang, H, Xue, X, Shen, W, Zhao, Q, Rao, Z. | | 登録日 | 2006-05-20 | | 公開日 | 2007-04-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction
J.Mol.Biol., 366, 2007
|
|
2HOB
 
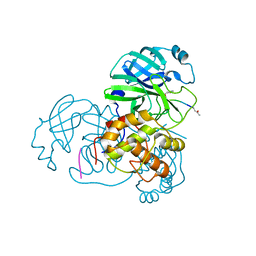 | | Crystal structure of SARS-CoV main protease with authentic N and C-termini in complex with a Michael acceptor N3 | | 分子名称: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, Replicase polyprotein 1ab | | 著者 | Xue, X, Yang, H, Shen, W, Zhao, Q, Li, J, Rao, Z. | | 登録日 | 2006-07-14 | | 公開日 | 2007-04-03 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction
J.Mol.Biol., 366, 2007
|
|
3QK0
 
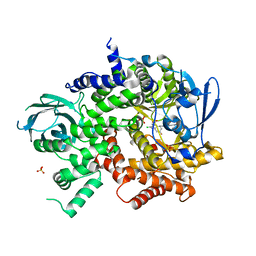 | | Crystal structure of PI3K-gamma in complex with benzothiazole 82 | | 分子名称: | N-[6-(6-chloro-5-{[(4-fluorophenyl)sulfonyl]amino}pyridin-3-yl)-1,3-benzothiazol-2-yl]acetamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | 著者 | Whittington, D.A, Tang, J, Yakowec, P. | | 登録日 | 2011-01-31 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Discovery and Optimization of a Series of Benzothiazole Phosphoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QJZ
 
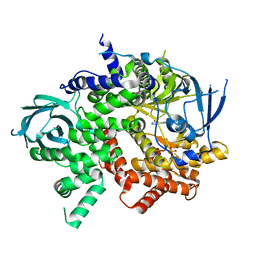 | | Crystal structure of PI3K-gamma in complex with benzothiazole 1 | | 分子名称: | N-{6-[2-(methylsulfanyl)pyrimidin-4-yl]-1,3-benzothiazol-2-yl}acetamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | 著者 | Whittington, D.A, Tang, J, Yakowec, P. | | 登録日 | 2011-01-31 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Discovery and Optimization of a Series of Benzothiazole Phosphoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors.
J.Med.Chem., 54, 2011
|
|
6M03
 
 | | The crystal structure of COVID-19 main protease in apo form | | 分子名称: | 3C-like proteinase | | 著者 | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Rao, Z. | | 登録日 | 2020-02-19 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5F9P
 
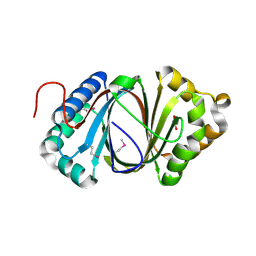 | | Crystal structure study of anthrone oxidase-like protein | | 分子名称: | Anthrone oxidase-like protein, GLYCEROL | | 著者 | Gao, X, Wu, D, Fan, K, Liu, Z.-J. | | 登録日 | 2015-12-10 | | 公開日 | 2016-12-14 | | 最終更新日 | 2018-07-18 | | 実験手法 | X-RAY DIFFRACTION (2.078 Å) | | 主引用文献 | Structure and Function of a C-C Bond Cleaving Oxygenase in Atypical Angucycline Biosynthesis
ACS Chem. Biol., 12, 2017
|
|
8W1N
 
 | |
5Y6N
 
 | | Zika virus helicase in complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Helicase domain from Genome polyprotein, MANGANESE (II) ION | | 著者 | Yang, X.Y, Chen, C, Tian, H.L, Chi, H, Mu, Z.Y, Zhang, T.Q, Yang, K.L, Zhao, Q, Liu, X.H, Wang, Z.F, Ji, X.Y, Yang, H.T. | | 登録日 | 2017-08-12 | | 公開日 | 2018-07-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.571 Å) | | 主引用文献 | Mechanism of ATP hydrolysis by the Zika virus helicase.
FASEB J., 32, 2018
|
|
8H3G
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166V Mutant in Complex with Inhibitor Enstrelvir | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, GLYCEROL | | 著者 | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3L
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (T21I and E166V) in Complex with Inhibitor Enstrelvir | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Enstrelvir | | 分子名称: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ... | | 著者 | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
5Y6M
 
 | | Zika virus helicase in complex with ADP-AlF3 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase domain from Genome polyprotein, ... | | 著者 | Yang, X.Y, Chen, C, Tian, H.L, Chi, H, Mu, Z.Y, Zhang, T.Q, Yang, K.L, Zhao, Q, Liu, X.H, Wang, Z.F, Ji, X.Y, Yang, H.T. | | 登録日 | 2017-08-12 | | 公開日 | 2018-07-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | Mechanism of ATP hydrolysis by the Zika virus helicase.
FASEB J., 32, 2018
|
|
