3M89
 
 | | Structure of TubZ-GTP-g-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, FtsZ/tubulin-related protein | | Authors: | Ni, L, Xu, W, Schumacher, M.A. | | Deposit date: | 2010-03-17 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | From the Cover: Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MKY
 
 | | Structure of SopB(155-323)-18mer DNA complex, I23 form | | Descriptor: | DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB, SULFATE ION | | Authors: | Schumacher, M.A, Piro, K, Xu, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
3Q7J
 
 | | Engineered Thermoplasma Acidophilum F3 factor mimics human aminopeptidase N (APN) as a target for anticancer drug development | | Descriptor: | L-phenylalanyl-N6-[(benzyloxy)carbonyl]-N1-hydroxy-L-lysinamide, Tricorn protease-interacting factor F3, ZINC ION | | Authors: | Su, J, Wang, Q, Feng, J, Zhang, C, Zhu, D, We, T, Xu, W, Gu, L. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Engineered Thermoplasma acidophilum factor F3 mimics human aminopeptidase N (APN) as a target for anticancer drug development
Bioorg.Med.Chem., 19, 2011
|
|
3S8V
 
 | | Crystal structure of LRP6-Dkk1 complex | | Descriptor: | Dickkopf-related protein 1, Low-density lipoprotein receptor-related protein 6 | | Authors: | Cheng, Z, Xu, W. | | Deposit date: | 2011-05-31 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the extracellular domain of LRP6 and its complex with DKK1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3S94
 
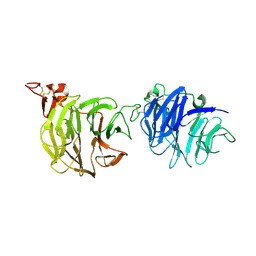 | | Crystal structure of LRP6-E1E2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Low-density lipoprotein receptor-related protein 6 | | Authors: | Cheng, Z, Xu, W. | | Deposit date: | 2011-05-31 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the extracellular domain of LRP6 and its complex with DKK1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3S8Z
 
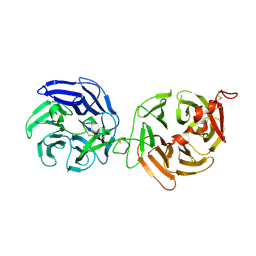 | | Crystal structure of LRP6-E3E4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Low-density lipoprotein receptor-related protein 6 | | Authors: | Cheng, Z, Xu, W. | | Deposit date: | 2011-05-31 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the extracellular domain of LRP6 and its complex with DKK1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3UTM
 
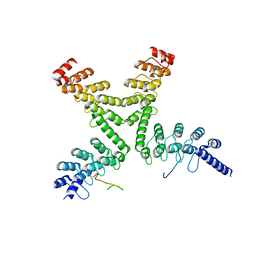 | | Crystal structure of a mouse Tankyrase-Axin complex | | Descriptor: | Axin-1, Tankyrase-1 | | Authors: | Cheng, Z, Morrone, S, Xu, W. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a Tankyrase-Axin complex and its implications for Axin turnover and Tankyrase substrate recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3V3L
 
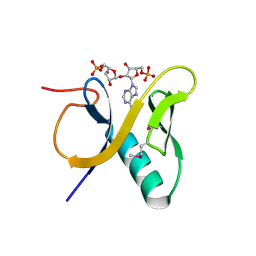 | | Crystal structure of human RNF146 WWE domain in complex with iso-ADPRibose | | Descriptor: | 2'-O-(5-O-phosphono-alpha-D-ribofuranosyl)adenosine 5'-(dihydrogen phosphate), E3 ubiquitin-protein ligase RNF146 | | Authors: | Wang, Z, Cheng, Z, Xu, W. | | Deposit date: | 2011-12-13 | | Release date: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination.
Genes Dev., 26, 2012
|
|
4EFE
 
 | | crystal structure of DNA ligase | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, SULFATE ION, ... | | Authors: | Wei, Y, Wang, T, Charifson, P, Xu, W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of DNA ligase
To be Published
|
|
4EFB
 
 | | Crystal structure of DNA ligase | | Descriptor: | 4-amino-2-(cyclopentyloxy)-6-{[(1R,2S)-2-hydroxycyclopentyl]oxy}pyrimidine-5-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Wei, Y, Wang, T, Charifson, P, Xu, W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of DNA ligase
To be Published
|
|
4FC2
 
 | |
4IBG
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | GLYCEROL, PHOSPHATE ION, Polymerase cofactor VP35, ... | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.413 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBD
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 5-[(2R)-3-benzoyl-2-(4-bromothiophen-2-yl)-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl]-2-methylbenzoic acid, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBB
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | Polymerase cofactor VP35, {4-[(5R)-3-hydroxy-2-oxo-4-(thiophen-2-ylcarbonyl)-5-(2,4,5-trimethylphenyl)-2,5-dihydro-1H-pyrrol-1-yl]phenyl}acetic acid | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-08 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBI
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 3-{(2S)-2-(7-chloro-1,3-benzodioxol-5-yl)-4-hydroxy-5-oxo-3-[3-(trifluoromethyl)benzoyl]-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.473 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBF
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | (4-{(2R)-2-(4-bromothiophen-2-yl)-3-[(5-chlorothiophen-2-yl)carbonyl]-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl}phenyl)acetic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBJ
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 3-{(5S)-3-hydroxy-2-oxo-4-[3-(trifluoromethyl)benzoyl]-5-[3-(trifluoromethyl)phenyl]-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBE
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 5-[(2R)-3-benzoyl-2-(4-bromothiophen-2-yl)-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl]-2-chlorobenzoic acid, GLYCEROL, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
6KHX
 
 | | Crystal structure of Prx from Akkermansia muciniphila | | Descriptor: | CALCIUM ION, Peroxiredoxin | | Authors: | Li, M, Wang, J, Xu, W, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of Akkermansia muciniphila peroxiredoxin reveals a novel regulatory mechanism of typical 2-Cys Prxs by a distinct loop.
Febs Lett., 594, 2020
|
|
6LND
 
 | | Crystal structure of transposition protein TniQ | | Descriptor: | ZINC ION, transposition protein TniQ | | Authors: | Wang, B, Xu, W, Yang, H. | | Deposit date: | 2019-12-28 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis of a Tn7-like transposase recruitment and DNA loading to CRISPR-Cas surveillance complex.
Cell Res., 30, 2020
|
|
6LNB
 
 | | CryoEM structure of Cascade-TniQ-dsDNA complex | | Descriptor: | CRISPR RNA (60-MER), CRISPR-associated protein Cas6, CRISPR-associated protein Cas7, ... | | Authors: | Wang, B, Xu, W, Yang, H. | | Deposit date: | 2019-12-28 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of a Tn7-like transposase recruitment and DNA loading to CRISPR-Cas surveillance complex.
Cell Res., 30, 2020
|
|
6LNC
 
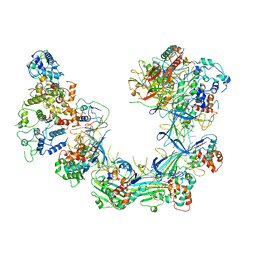 | | CryoEM structure of Cascade-TniQ complex | | Descriptor: | CRISPR RNA (60-MER), CRISPR-associated protein Cas6, CRISPR-associated protein Cas7, ... | | Authors: | Wang, B, Xu, W, Yang, H. | | Deposit date: | 2019-12-28 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of a Tn7-like transposase recruitment and DNA loading to CRISPR-Cas surveillance complex.
Cell Res., 30, 2020
|
|
5WZZ
 
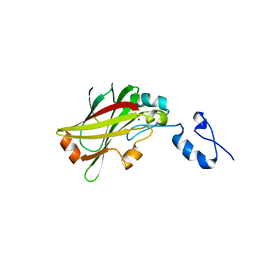 | | The SIAH E3 ubiquitin ligases promote Wnt/ beta-catenin signaling through mediating Wnt-induced Axin degradation | | Descriptor: | Axin-1, E3 ubiquitin-protein ligase SIAH1, ZINC ION | | Authors: | Ji, L, Jiang, B, Jiang, X, Charlat, O, Chen, A, Mickanin, C, Bauer, A, Xu, W, Yan, X.-X, Cong, F. | | Deposit date: | 2017-01-19 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | The SIAH E3 ubiquitin ligases promote Wnt/ beta-catenin signaling through mediating Wnt-induced Axin degradation
Genes Dev., 31, 2017
|
|
6BMG
 
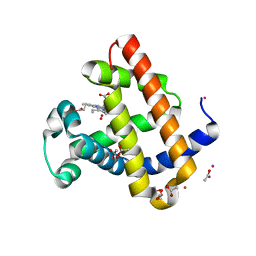 | | Structure of Recombinant Dwarf Sperm Whale Myoglobin (Oxy) | | Descriptor: | ACETATE ION, CADMIUM ION, Myoglobin, ... | | Authors: | Samuel, P.P, Miller, M.D, Xu, W, Alvarado, S, Phillips Jr, G.N, Olson, J.S. | | Deposit date: | 2017-11-14 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure of Recombinant Dwarf Sperm Whale Myoglobin (Oxy)
To Be Published
|
|
6BUI
 
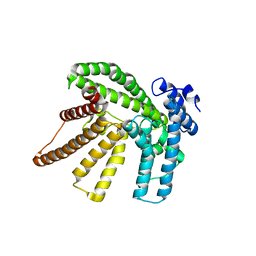 | |
