7JPY
 
 | |
7JQ4
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI7 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-phenylalaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JQ0
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI3 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JQ1
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI4 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl]propan-2-yl}-L-phenylalaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JPZ
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI1 | | Descriptor: | (phenylmethyl) N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate, 3C-like proteinase | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JQ5
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI8 | | Descriptor: | 3C-like proteinase, N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JQ3
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI6 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
6KKE
 
 | | THRb mutation with a novel agonist | | Descriptor: | 2-[(1-methyl-4-oxidanyl-7-phenoxy-isoquinolin-3-yl)carbonylamino]ethanoic acid, SRC2-3, Thyroid hormone receptor beta | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2019-07-25 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.577 Å) | | Cite: | Revealing a Mutant-Induced Receptor Allosteric Mechanism for the Thyroid Hormone Resistance.
Iscience, 20, 2019
|
|
6KKB
 
 | | A novel agonist of THRb | | Descriptor: | 2-[(1-methyl-4-oxidanyl-7-phenoxy-isoquinolin-3-yl)carbonylamino]ethanoic acid, SRC2-3, Thyroid hormone receptor beta | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2019-07-24 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Revealing a Mutant-Induced Receptor Allosteric Mechanism for the Thyroid Hormone Resistance.
Iscience, 20, 2019
|
|
6KNU
 
 | | THRb mutation with a novel agonist | | Descriptor: | 2-[(1-methyl-4-oxidanyl-7-phenoxy-isoquinolin-3-yl)carbonylamino]ethanoic acid, Nuclear receptor coactivator 2, Thyroid hormone receptor beta | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2019-08-07 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Revealing a Mutant-Induced Receptor Allosteric Mechanism for the Thyroid Hormone Resistance.
Iscience, 20, 2019
|
|
7JS8
 
 | | STRUCTURE OF HUMAN HDAC2 IN COMPLEX WITH AN ETHYL KETONE INHIBITOR CONTAINING A SPIRO-BICYCLIC GROUP (COMPOUND 22) | | Descriptor: | (1S)-N-{(1S)-7,7-dihydroxy-1-[4-(2-methylquinolin-6-yl)-1H-imidazol-2-yl]nonyl}-6-methyl-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2020-08-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.634 Å) | | Cite: | Discovery of Ethyl Ketone-Based Highly Selective HDACs 1, 2, 3 Inhibitors for HIV Latency Reactivation with Minimum Cellular Potency Serum Shift and Reduced hERG Activity.
J.Med.Chem., 64, 2021
|
|
6M05
 
 | | Trimolecular G-quadruplex | | Descriptor: | DNA (5'-D(*GP*TP*TP*AP*GP*G)-3') | | Authors: | Jing, H.T, Fu, W.Q, Zhang, N. | | Deposit date: | 2020-02-20 | | Release date: | 2021-01-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structural study on the self-trimerization of d(GTTAGG) into a dynamic trimolecular G-quadruplex assembly preferentially in Na+ solution with a moderate K+ tolerance.
Nucleic Acids Res., 49, 2021
|
|
8STZ
 
 | |
8STY
 
 | |
2Q7M
 
 | | Crystal structure of human FLAP with MK-591 | | Descriptor: | 3-[3-(TERT-BUTYLTHIO)-1-(4-CHLOROBENZYL)-5-(QUINOLIN-2-YLMETHOXY)-1H-INDOL-2-YL]-2,2-DIMETHYLPROPANOIC ACID, Arachidonate 5-lipoxygenase-activating protein | | Authors: | Ferguson, A.D. | | Deposit date: | 2007-06-07 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein.
Science, 317, 2007
|
|
2Q7R
 
 | | Crystal structure of human FLAP with an iodinated analog of MK-591 | | Descriptor: | 3-[3-(3,3-DIMETHYLBUTANOYL)-1-(4-IODOBENZYL)-5-(QUINOLIN-2-YLMETHOXY)-1H-INDOL-2-YL]-2,2-DIMETHYLPROPANOIC ACID, Arachidonate 5-lipoxygenase-activating protein | | Authors: | Ferguson, A.D. | | Deposit date: | 2007-06-07 | | Release date: | 2007-08-21 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein.
Science, 317, 2007
|
|
8W41
 
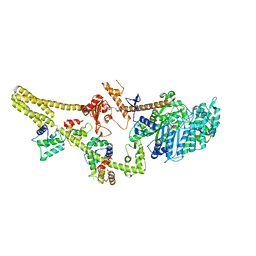 | | Cryo-EM structure of Myosin VI in the autoinhibited state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Calmodulin-1, ... | | Authors: | Niu, F, Wei, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Autoinhibition and activation of myosin VI revealed by its cryo-EM structure.
Nat Commun, 15, 2024
|
|
8GWO
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWI
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWB
 
 | | SARS-CoV-2 E-RTC complex with RNA-nsp9 | | Descriptor: | Helicase, MANGANESE (II) ION, Non-structural protein 7, ... | | Authors: | Yan, L.M, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-16 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWG
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWN
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibitor of AT-527 | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWF
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | Authors: | Yan, L.Y, Huang, Y.C, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWE
 
 | | SARS-CoV-2 E-RTC complex with RNA-nsp9 and GMPPNP | | Descriptor: | Helicase nsp13, MAGNESIUM ION, Non-structural protein 8, ... | | Authors: | Yan, L.M, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-16 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GW1
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | Helicase, MANGANESE (II) ION, Non-structural protein 7, ... | | Authors: | Yan, L, Rao, Z, Lou, Z. | | Deposit date: | 2022-09-16 | | Release date: | 2023-10-25 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
