3TEF
 
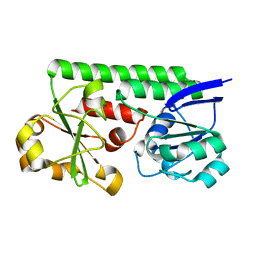 | | Crystal Structure of the Periplasmic Catecholate-Siderophore Binding Protein VctP from Vibrio Cholerae | | Descriptor: | Iron(III) ABC transporter, periplasmic iron-compound-binding protein | | Authors: | Liu, X, Wang, Z, Liu, S, Li, N, Chen, Y, Zhu, C, Zhu, D, Wei, T, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-08-13 | | Release date: | 2012-08-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of periplasmic catecholate-siderophore binding protein VctP from Vibrio cholerae at 1.7 A resolution
Febs Lett., 586, 2012
|
|
3TB4
 
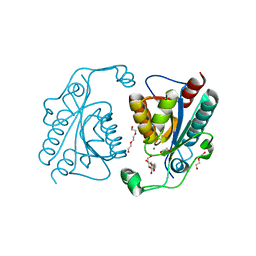 | | Crystal structure of the ISC domain of VibB | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liu, S, Zhang, C, Niu, B, Li, N, Liu, M, Wei, T, Zhu, D, Xu, S, Gu, L. | | Deposit date: | 2011-08-05 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insight into the ISC domain of VibB from Vibrio cholerae at atomic resolution: a snapshot just before the enzymatic reaction
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5GK9
 
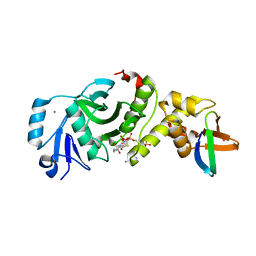 | | Crystal structure of human HBO1 in complex with BRPF2 | | Descriptor: | ACETYL COENZYME *A, BRD1 protein, Histone acetyltransferase KAT7, ... | | Authors: | Tao, Y, Zhu, J, Xu, S, Ding, J. | | Deposit date: | 2016-07-04 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and mechanistic insights into regulation of HBO1 histone acetyltransferase activity by BRPF2.
Nucleic Acids Res., 45, 2017
|
|
3O72
 
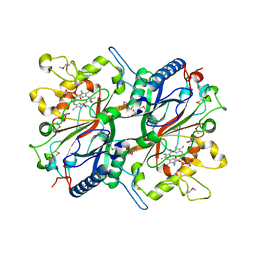 | | Crystal structure of EfeB in complex with heme | | Descriptor: | OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, Redox component of a tripartite ferrous iron transporter | | Authors: | Liu, X, Du, Q, Wang, Z, Zhu, D, Huang, Y, Li, N, Xu, S, Gu, L. | | Deposit date: | 2010-07-30 | | Release date: | 2011-03-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and biochemical features of EfeB/YcdB from Escherichia coli O157: ASP235 plays divergent roles in different enzyme-catalyzed processes
J.Biol.Chem., 286, 2011
|
|
3R5S
 
 | | Crystal structure of apo-ViuP | | Descriptor: | Ferric vibriobactin ABC transporter, periplasmic ferric vibriobactin-binding protein | | Authors: | Li, N, Zhang, C, Li, B, Liu, X, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-03-19 | | Release date: | 2012-02-08 | | Last modified: | 2012-05-30 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Unique iron coordination in iron-chelating molecule vibriobactin helps Vibrio cholerae evade mammalian siderocalin-mediated immune response.
J.Biol.Chem., 287, 2012
|
|
3R5T
 
 | | Crystal structure of holo-ViuP | | Descriptor: | (4S,5R)-N-{3-[(2,3-dihydroxybenzoyl)amino]propyl}-2-(2,3-dihydroxyphenyl)-N-[3-({[(4S,5R)-2-(2,3-dihydroxyphenyl)-5-met hyl-4,5-dihydro-1,3-oxazol-4-yl]carbonyl}amino)propyl]-5-methyl-4,5-dihydro-1,3-oxazole-4-carboxamide, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Li, N, Zhang, C, Li, B, Liu, X, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-03-19 | | Release date: | 2012-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Unique iron coordination in iron-chelating molecule vibriobactin helps Vibrio cholerae evade mammalian siderocalin-mediated immune response.
J.Biol.Chem., 287, 2012
|
|
3SFV
 
 | | Crystal structure of the GDP-bound Rab1a S25N mutant in complex with the coiled-coil domain of LidA from Legionella pneumophila | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, LidA protein, substrate of the Dot/Icm system, ... | | Authors: | Yin, K, Lu, D, Zhu, D, Zhang, H, Li, B, Xu, S, Gu, L. | | Deposit date: | 2011-06-14 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Insights into a Unique Legionella pneumophila Effector LidA Recognizing Both GDP and GTP Bound Rab1 in Their Active State
Plos Pathog., 8, 2012
|
|
3REE
 
 | | Crystal structure of mitoNEET | | Descriptor: | CDGSH iron-sulfur domain-containing protein 1, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Funk, M.O, Arif, W, Xu, S, Mueser, T.C. | | Deposit date: | 2011-04-04 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Complexes of the Outer Mitochondrial Membrane Protein MitoNEET with Resveratrol-3-Sulfate.
Biochemistry, 50, 2011
|
|
3TGV
 
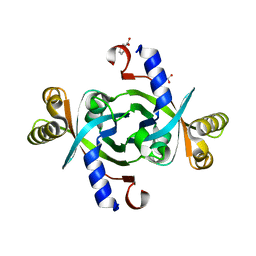 | | Crystal structure of HutZ,the heme storsge protein from Vibrio cholerae | | Descriptor: | BENZOIC ACID, Heme-binding protein HutZ | | Authors: | Liu, X, Gong, J, Wang, Z, Du, Q, Wei, T, Zhu, D, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-08-17 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystal structure of HutZ,the heme storsge protein from Vibrio cholerae
To be Published
|
|
2KB8
 
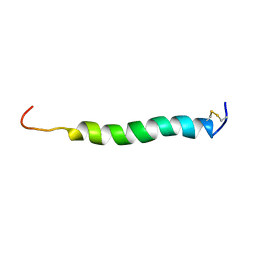 | |
4HKF
 
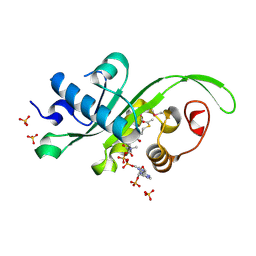 | | Crystal structure of Danio rerio MEC-17 catalytic domain in complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Alpha-tubulin N-acetyltransferase, SULFATE ION | | Authors: | Li, W, Zhong, C, Sun, B, Xu, S, Zhang, T, Ding, J. | | Deposit date: | 2012-10-15 | | Release date: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of the acetyltransferase activity of MEC-17 towards alpha- tubulin
Cell Res., 22, 2012
|
|
4F5Y
 
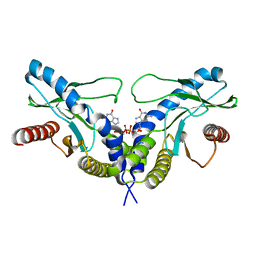 | | Crystal structure of human STING CTD complex with C-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Transmembrane protein 173 | | Authors: | Gu, L, Shang, G, Zhu, D, Li, N, Zhang, J, Zhu, C, Lu, D, Liu, C, Yu, Q, Zhao, Y, Xu, S. | | Deposit date: | 2012-05-13 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP
Nat.Struct.Mol.Biol., 19, 2012
|
|
4F5W
 
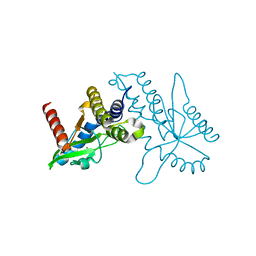 | | Crystal structure of ligand free human STING CTD | | Descriptor: | CALCIUM ION, Transmembrane protein 173 | | Authors: | Gu, L, Shang, G, Zhu, D, Li, N, Zhang, J, Zhu, C, Lu, D, Liu, C, Yu, Q, Zhao, Y, Xu, S. | | Deposit date: | 2012-05-13 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP
Nat.Struct.Mol.Biol., 19, 2012
|
|
5VQF
 
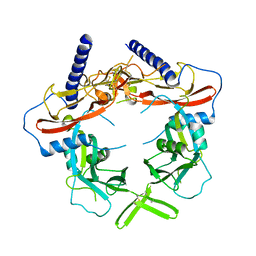 | | Crystal Structure of pro-TGF-beta 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Transforming growth factor beta-1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zhao, B, Xu, S, Dong, X, Lu, C, Springer, T.A. | | Deposit date: | 2017-05-08 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Prodomain-growth factor swapping in the structure of pro-TGF-beta 1.
J. Biol. Chem., 293, 2018
|
|
5VQP
 
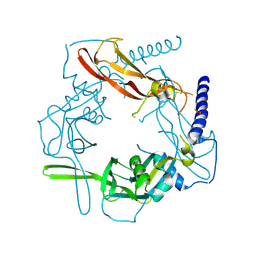 | | Crystal structure of human pro-TGF-beta1 | | Descriptor: | Transforming growth factor beta-1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zhao, B, Xu, S, Dong, X, Lu, C, Springer, T.A. | | Deposit date: | 2017-05-09 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Prodomain-growth factor swapping in the structure of pro-TGF-beta 1.
J. Biol. Chem., 293, 2018
|
|
7DKG
 
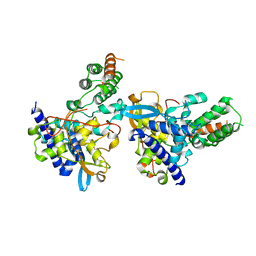 | | Influenza H5N1 nucleoprotein (truncated) in complex with nucleotides | | Descriptor: | Nucleoprotein, RNA (5'-R(P*(OMU)P*(OMU)P*(OMU))-3') | | Authors: | Tang, Y.S, Xu, S, Chen, Y.W, Wang, J.H, Shaw, P.C. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of influenza nucleoprotein complexed with nucleic acid provide insights into the mechanism of RNA interaction.
Nucleic Acids Res., 49, 2021
|
|
7EO6
 
 | | X-ray structure analysis of xylanase | | Descriptor: | Endo-1,4-beta-xylanase, IODIDE ION | | Authors: | Wan, Q, Yi, Y, Xu, S. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization and structural analysis of a thermophilic GH11 xylanase from compost metatranscriptome.
Appl.Microbiol.Biotechnol., 105, 2021
|
|
7EO3
 
 | | X-ray structure analysis of beita-1,3-glucanase | | Descriptor: | 1,3-beta-glucanase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION | | Authors: | Wan, Q, Feng, J, Xu, S. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.141 Å) | | Cite: | Identification and structural analysis of a thermophilic beta-1,3-glucanase from compost
Bioresour Bioprocess, 8, 2021
|
|
7DXP
 
 | | Influenza H5N1 nucleoprotein in complex with nucleotides | | Descriptor: | 1,2-ETHANEDIOL, Nucleoprotein, RNA (5'-R(P*(OMU)P*(OMU)P*(OMU)P*(OMU))-3') | | Authors: | Tang, Y.S, Xu, S, Chen, Y.W, Wang, J.H, Shaw, P.C. | | Deposit date: | 2021-01-19 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of influenza nucleoprotein complexed with nucleic acid provide insights into the mechanism of RNA interaction.
Nucleic Acids Res., 49, 2021
|
|
6WNH
 
 | | Menin bound to inhibitor M-808 | | Descriptor: | Menin, methyl [(1S,2R)-2-{(1S)-2-(azetidin-1-yl)-1-(3-fluorophenyl)-1-[1-({1-[4-({1-[4-(piperidin-1-yl)butanoyl]azetidin-3-yl}sulfonyl)phenyl]azetidin-3-yl}methyl)piperidin-4-yl]ethyl}cyclopentyl]carbamate, praseodymium triacetate | | Authors: | Stuckey, J.A. | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of M-808 as a Highly Potent, Covalent, Small-Molecule Inhibitor of the Menin-MLL Interaction with StrongIn VivoAntitumor Activity.
J.Med.Chem., 63, 2020
|
|
6B41
 
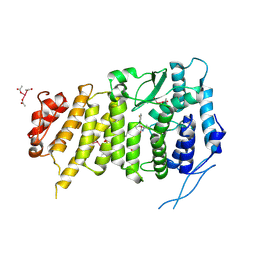 | | Menin bound to M-525 | | Descriptor: | Menin, methyl {(1S,2R)-2-[(S)-cyano[1-({1-[4-({1-[4-(dimethylamino)butanoyl]azetidin-3-yl}sulfonyl)phenyl]azetidin-3-yl}methyl)piperidin-4-yl](3-fluorophenyl)methyl]cyclopentyl}carbamate, praseodymium triacetate | | Authors: | Stuckey, J.A. | | Deposit date: | 2017-09-25 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Design of the First-in-Class, Highly Potent Irreversible Inhibitor Targeting the Menin-MLL Protein-Protein Interaction.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8TQP
 
 | | HIV-CA Disulfide linked Hexamer bound to Quinazolin-4-one Scaffold inhibitor | | Descriptor: | 2-[4-(4-aminobenzene-1-sulfonyl)-2-oxopiperazin-1-yl]-N-{(1R)-2-(3,5-difluorophenyl)-1-[3-(4-methoxyphenyl)-4-oxo-3,4-dihydroquinazolin-2-yl]ethyl}acetamide, Gag polyprotein | | Authors: | Goldstone, D.C, Barnett, M.J, Taka, J.R.H. | | Deposit date: | 2023-08-08 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery, Crystallographic Studies, and Mechanistic Investigations of Novel Phenylalanine Derivatives Bearing a Quinazolin-4-one Scaffold as Potent HIV Capsid Modulators.
J.Med.Chem., 66, 2023
|
|
8TOV
 
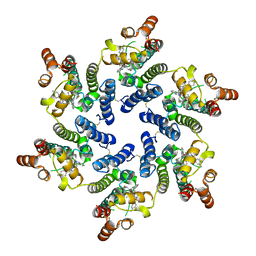 | | HIV-CA Disulfide linked Hexamer bound to Quinazolin-4-one Scaffold inhibitor | | Descriptor: | 2-[4-(4-aminobenzene-1-sulfonyl)-2-oxopiperazin-1-yl]-N-[(1R)-2-(3,5-difluorophenyl)-1-{3-[4-(morpholine-4-sulfonyl)phenyl]-4-oxo-3,4-dihydroquinazolin-2-yl}ethyl]acetamide, Matrix protein p17 | | Authors: | Goldstone, D.C, Barnett, M.J, Taka, J.R.H. | | Deposit date: | 2023-08-04 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery, Crystallographic Studies, and Mechanistic Investigations of Novel Phenylalanine Derivatives Bearing a Quinazolin-4-one Scaffold as Potent HIV Capsid Modulators.
J.Med.Chem., 66, 2023
|
|
7N02
 
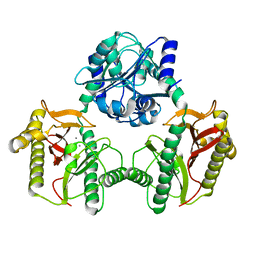 | |
6XMR
 
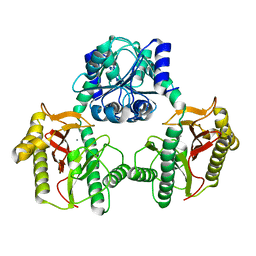 | |
