1MQ9
 
 | | Crystal structure of high affinity alphaL I domain with ligand mimetic crystal contact | | Descriptor: | Integrin alpha-L, MANGANESE (II) ION | | Authors: | Shimaoka, M, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, McCormack, A, Zhang, R, Joachimiak, A, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
5XIV
 
 | | Beta-Ginkgotides: Hyperdisulfide-constrained peptides from Ginkgo biloba | | Descriptor: | beta-ginkgotide, beta-gB1 | | Authors: | Wong, K.H, Tan, W.L, Xiao, T, Tam, J.P. | | Deposit date: | 2017-04-27 | | Release date: | 2017-05-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | beta-Ginkgotides: Hyperdisulfide-constrained peptides from Ginkgo biloba.
Sci Rep, 7, 2017
|
|
3MES
 
 | | Crystal structure of choline kinase from Cryptosporidium parvum Iowa II, cgd3_2030 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Choline kinase, DECAMETHONIUM ION, ... | | Authors: | Qiu, W, Wernimont, A, Hills, T, Lew, J, Artz, J.D, Xiao, T, Allali-Hassani, A, Vedadi, M, Kozieradzki, I, Cossar, D, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Hui, R, Ma, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of choline kinase from Cryptosporidium parvum Iowa II, cgd3_2030
TO BE PUBLISHED
|
|
3C5I
 
 | | Crystal structure of Plasmodium knowlesi choline kinase, PKH_134520 | | Descriptor: | CALCIUM ION, CHOLINE ION, Choline kinase, ... | | Authors: | Wernimont, A.K, Hills, T, Lew, J, Wasney, G, Senesterra, G, Kozieradzki, I, Cossar, D, Vedadi, M, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Plasmodium knowlesi choline kinase, PKH_134520.
To be Published
|
|
3FI8
 
 | | Crystal structure of choline kinase from Plasmodium Falciparum, PF14_0020 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Choline kinase, MAGNESIUM ION, ... | | Authors: | Wernimont, A.K, Pizarro, J.C, Artz, J.D, Amaya, M.F, Xiao, T, Lew, J, Wasney, G, Senesterra, G, Kozieradzki, I, Cossar, D, Vedadi, M, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-11 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of choline kinase from Plasmodium Falciparum, PF14_0020
TO BE PUBLISHED
|
|
3KLW
 
 | | Crystal structure of primosomal replication protein n from Bordetella pertussis. northeast structural genomics consortium target ber132. | | Descriptor: | Primosomal replication protein n | | Authors: | Kuzin, A.P, Neely, H, Vorobiev, S.M, Seetharaman, J, Forouhar, F, Wang, H, Janjua, H, Maglaqui, M, Xiao, T, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-09 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of primosomal replication protein n from Bordetella pertussis. northeast structural genomics consortium target ber132.
To be Published
|
|
2POY
 
 | | Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_4120 in complex with cyclosporin A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Wernimont, A.K, Lew, J, Hills, T, Kozieradzki, I, Lin, Y.H, Hassanali, A, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-27 | | Release date: | 2007-05-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cryptosporidium Parvum Cyclophilin Type Peptidyl-Prolyl Cis-Trans Isomerase Cgd2_4120 in Complex with Cyclosporin A.
To be Published
|
|
2PLU
 
 | | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_4120 | | Descriptor: | 20k cyclophilin, putative | | Authors: | Wernimont, A.K, Lew, J, Hills, T, Kozieradzki, I, Lin, Y.H, Hassanali, A, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_4120.
To be Published
|
|
2QG7
 
 | | Plasmodium vivax ethanolamine kinase Pv091845 | | Descriptor: | SULFATE ION, ethanolamine kinase Pv091845 | | Authors: | Lunin, V.V, Wernimont, A.K, Mulichak, A, Lew, J, Wasney, G, Senisterra, G, Kozieradzki, I, Vedadi, M, Bochkarev, A, Arrowsmith, C.H, Sundstrom, M, Weigelt, J, Edwards, A.E, Hui, R, Hills, T, Artz, J, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Plasmodium vivax ethanolamine kinase Pv091845
TO BE PUBLISHED
|
|
3B6N
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase PV003920 from Plasmodium vivax | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ZINC ION | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase PV003920 from Plasmodium vivax.
To be Published
|
|
3BO7
 
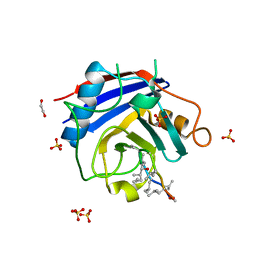 | | Crystal structure of Toxoplasma gondii peptidyl-prolyl cis-trans isomerase, 541.m00136 | | Descriptor: | 1,2-ETHANEDIOL, CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE CYCLOPHILIN-TYPE, ... | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-17 | | Release date: | 2008-02-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Toxoplasma Gondii Peptidyl-Prolyl Cis-Trans Isomerase, 541.M00136.
To be Published
|
|
3DFA
 
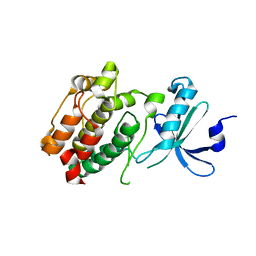 | | Crystal structure of kinase domain of calcium-dependent protein kinase cgd3_920 from Cryptosporidium parvum | | Descriptor: | Calcium-dependent protein kinase cgd3_920 | | Authors: | Wernimont, A.K, Lew, J, Lin, Y.H, Hassanali, A, Khuu, C, Alam, Z, Kozieradzki, I, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Wilkstrom, M, Edwards, A.M, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-11 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of kinase domain of calcium-dependent protein kinase cgd3_920 from Cryptosporidium parvum.
To be Published
|
|
3FCU
 
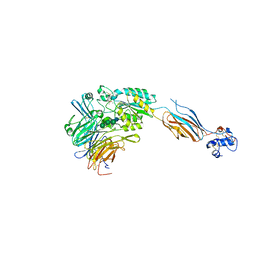 | | Structure of headpiece of integrin aIIBb3 in open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Zhu, J, Luo, B.-H, Xiao, T, Zhang, C, Nishida, N, Springer, T.A. | | Deposit date: | 2008-11-22 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces.
Mol.Cell, 32, 2008
|
|
3FCS
 
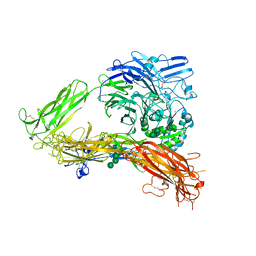 | | Structure of complete ectodomain of integrin aIIBb3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhu, J, Luo, B.-H, Xiao, T, Zhang, C, Nishida, N, Springer, T.A. | | Deposit date: | 2008-11-22 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces.
Mol.Cell, 32, 2008
|
|
2R77
 
 | | Crystal structure of phosphatidylethanolamine-binding protein, pfl0955c, from Plasmodium falciparum | | Descriptor: | Phosphatidylethanolamine-binding protein, putative | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Crombette, L, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-07 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of phosphatidylethanolamine-binding protein, pfl0955c, from Plasmodium falciparum.
To be Published
|
|
7MPH
 
 | | GRB2 SH2 Domain with Compound 7 | | Descriptor: | (4-{(10R,11E,14S,18S)-18-(2-amino-2-oxoethyl)-14-[(naphthalen-1-yl)methyl]-8,17,20-trioxo-7,16,19-triazaspiro[5.14]icos-11-en-10-yl}phenyl)acetic acid, 1,2-ETHANEDIOL, Growth factor receptor-bound protein 2, ... | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and structural characterization of a monocarboxylic inhibitor for GRB2 SH2 domain.
Bioorg.Med.Chem.Lett., 51, 2021
|
|
7KJ3
 
 | | SARS-CoV-2 Spike Glycoprotein with two ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KJ5
 
 | | SARS-CoV-2 Spike Glycoprotein, prefusion with one RBD up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KJ2
 
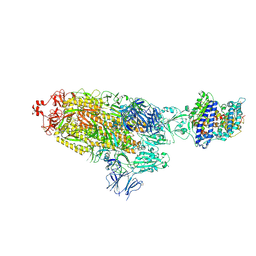 | | SARS-CoV-2 Spike Glycoprotein with one ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KJ4
 
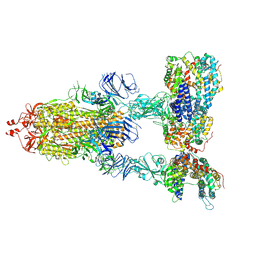 | | SARS-CoV-2 Spike Glycoprotein with three ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6L7K
 
 | | solution structure of hIFABP V60C/Y70C variant. | | Descriptor: | Fatty acid-binding protein, intestinal | | Authors: | Fan, J, Yang, D. | | Deposit date: | 2019-11-01 | | Release date: | 2020-11-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Ligand Entry into Fatty Acid Binding Protein via Local Unfolding Instead of Gap Widening.
Biophys.J., 118, 2020
|
|
5H1I
 
 | | NMR structure of TIBA, a chimera of SFTI | | Descriptor: | Bradykinin-trypsin inhibitor secondary loop chimera | | Authors: | Xiao, T, Tam, J.P. | | Deposit date: | 2016-10-10 | | Release date: | 2017-04-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | An Orally Active Bradykinin B1 Receptor Antagonist Engineered as a Bifunctional Chimera of Sunflower Trypsin Inhibitor.
J. Med. Chem., 60, 2017
|
|
5H1H
 
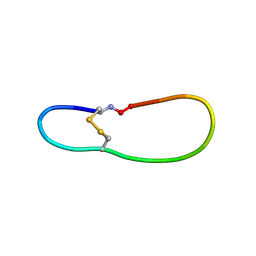 | | NMR structure of SLBA, a chimera of SFTI | | Descriptor: | Bradykinin-trypsin inhibitor secondary loop chimera | | Authors: | Xiao, T, Tam, J.P. | | Deposit date: | 2016-10-10 | | Release date: | 2017-04-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | An Orally Active Bradykinin B1 Receptor Antagonist Engineered as a Bifunctional Chimera of Sunflower Trypsin Inhibitor.
J. Med. Chem., 60, 2017
|
|
7F0T
 
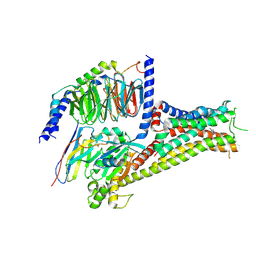 | |
7F1O
 
 | |
