4ZSE
 
 | |
3V43
 
 | | Crystal structure of MOZ | | Descriptor: | ACETATE ION, Histone H3.1, Histone acetyltransferase KAT6A, ... | | Authors: | Qiu, Y, Li, F. | | Deposit date: | 2011-12-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription
Genes Dev., 26, 2012
|
|
6RZ3
 
 | | Crystal structure of a complex between the DNA-binding domain of p53 and the carboxyl-terminal conserved region of iASPP | | Descriptor: | Cellular tumor antigen p53, RelA-associated inhibitor, ZINC ION | | Authors: | Chen, S, Ren, J, Jones, E.Y, Lu, X. | | Deposit date: | 2019-06-12 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.23 Å) | | Cite: | iASPP mediates p53 selectivity through a modular mechanism fine-tuning DNA recognition.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6XZ8
 
 | | Structure of aldosterone synthase (CYP11B2) in complex with N-[(1R)-1-[5-(6-chloro-1,1-dimethyl-3-oxo-isoindolin-2-yl)-3-pyridyl]ethyl]methanesulfonamide | | Descriptor: | Cytochrome P450 11B2, mitochondrial, HEME C, ... | | Authors: | Kuglstatter, A, Joseph, C, Benz, J. | | Deposit date: | 2020-02-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of 3-Pyridyl Isoindolin-1-one Derivatives as Potent, Selective, and Orally Active Aldosterone Synthase (CYP11B2) Inhibitors.
J.Med.Chem., 63, 2020
|
|
6XZ9
 
 | | Structure of aldosterone synthase (CYP11B2) in complex with 5-chloro-3,3-dimethyl-2-[5-[1-(1-methylpyrazole-4-carbonyl)azetidin-3-yl]oxy-3-pyridyl]isoindolin-1-one | | Descriptor: | 5-chloranyl-3,3-dimethyl-2-[5-[1-(1-methylpyrazol-4-yl)carbonylazetidin-3-yl]oxypyridin-3-yl]isoindol-1-one, Cytochrome P450 11B2, mitochondrial, ... | | Authors: | Kuglstatter, A, Joseph, C, Benz, J. | | Deposit date: | 2020-02-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Discovery of 3-Pyridyl Isoindolin-1-one Derivatives as Potent, Selective, and Orally Active Aldosterone Synthase (CYP11B2) Inhibitors.
J.Med.Chem., 63, 2020
|
|
8V1L
 
 | |
5DP4
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 3 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[(2S)-2-methyl-3-phenylpropanoyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP9
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 9 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-{[(cyclobutylmethyl)amino](oxo)acetyl}-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP8
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 8 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-{[(2-cyclopropylethyl)amino](oxo)acetyl}-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DPA
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 6 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-acetyl-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP5
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 4 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[(2R,5S)-5-amino-2-(4-fluorobenzyl)-6-methyl-4-oxoheptanoyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2016-04-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP7
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 5 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[N-(3-methylbutanoyl)-L-phenylalanyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP3
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 2 | | Descriptor: | 3C proteinase, ethyl (4S)-5-[(3S)-2-oxopyrrolidin-3-yl]-4-[(3-phenylpropanoyl)amino]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP6
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 7 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[N-(3-cyclopropylpropanoyl)-L-phenylalanyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
6LDZ
 
 | | Crystal structure of Rv0222 from Mycobacterium tuberculosis | | Descriptor: | Probable enoyl-CoA hydratase EchA1 (Enoyl hydrase) (Unsaturated acyl-CoA hydratase) (Crotonase) | | Authors: | Li, J, Ran, Y.J, Wang, L, Wu, J.H, Ge, B.X, Rao, Z.H. | | Deposit date: | 2019-11-23 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Host-mediated ubiquitination of a mycobacterial protein suppresses immunity.
Nature, 577, 2020
|
|
5GI4
 
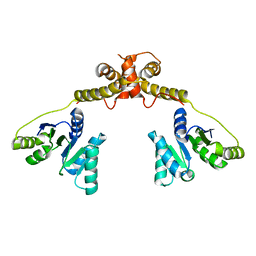 | | DEAD-box RNA helicase | | Descriptor: | ATP-dependent RNA helicase DeaD | | Authors: | Xu, L, Wang, L, Li, F, Wu, L, Shi, Y. | | Deposit date: | 2016-06-22 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
6URC
 
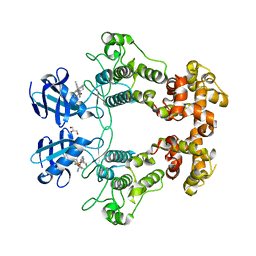 | | Crystal structure of IRE1a in complex with compound 18 | | Descriptor: | 2-chloro-N-(6-methyl-5-{[3-(2-{[(3S)-piperidin-3-yl]amino}pyrimidin-4-yl)pyridin-2-yl]oxy}naphthalen-1-yl)benzene-1-sulfonamide, GLYCEROL, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Wallweber, H.H, Wang, W. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Disruption of IRE1 alpha through its kinase domain attenuates multiple myeloma.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6CQD
 
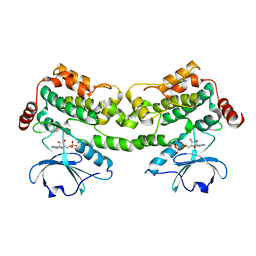 | | Crystal structure of HPK1 in complex with ATP analogue (AMPPNP) | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase kinase kinase kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wu, P, Lehoux, I, Franke, Y, Mortara, K, Wang, W. | | Deposit date: | 2018-03-14 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Hematopoietic Progenitor Kinase-1 Structure in a Domain-Swapped Dimer.
Structure, 27, 2019
|
|
6CQE
 
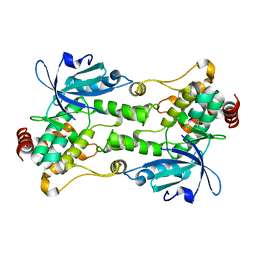 | | Crystal structure of HPK1 kinase domain S171A mutant | | Descriptor: | Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Wu, P, Lehoux, I, Mortara, K, Franke, Y, Wang, W. | | Deposit date: | 2018-03-15 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.886 Å) | | Cite: | Hematopoietic Progenitor Kinase-1 Structure in a Domain-Swapped Dimer.
Structure, 27, 2019
|
|
6CQF
 
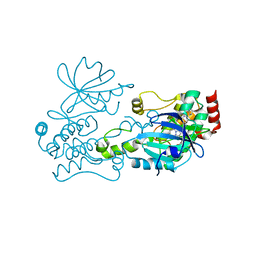 | | Crystal structure of HPK1 in complex an inhibitor G1858 | | Descriptor: | Mitogen-activated protein kinase kinase kinase kinase 1, N-{2-(3,3-difluoropyrrolidin-1-yl)-6-[(3R)-pyrrolidin-3-yl]pyrimidin-4-yl}-1-(propan-2-yl)-1H-pyrazolo[4,3-c]pyridin-6-amine | | Authors: | Wu, P, Lehoux, I, Mortara, K, Franke, Y, Chan, B.K, Wang, W. | | Deposit date: | 2018-03-15 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Hematopoietic Progenitor Kinase-1 Structure in a Domain-Swapped Dimer.
Structure, 27, 2019
|
|
3V53
 
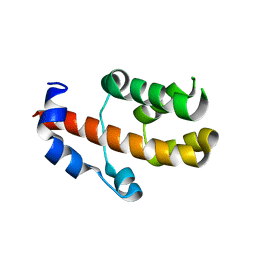 | | Crystal structure of human RBM25 | | Descriptor: | RNA-binding protein 25 | | Authors: | Gong, D.S. | | Deposit date: | 2011-12-16 | | Release date: | 2012-12-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and functional characterization of the human RBM25 PWI domain and its flanking basic region
Biochem.J., 450, 2013
|
|
7SL7
 
 | |
7SL1
 
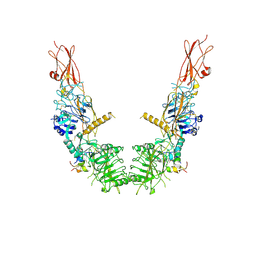 | |
7SL3
 
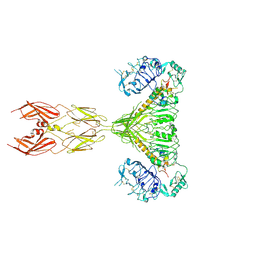 | |
7SL4
 
 | |
