8ETU
 
 | | Class2 of the INO80-Hexasome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETW
 
 | | Class3 of INO80-Hexasome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETS
 
 | | Class1 of the INO80-Hexasome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8EUJ
 
 | | Class2 of the INO80-Nucleosome complex | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | Deposit date: | 2022-10-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
2FJ4
 
 | |
2FJ5
 
 | |
2MVM
 
 | | Solution structure of eEF1Bdelta CAR domain | | Descriptor: | Elongation factor 1-delta | | Authors: | Wu, H, Feng, Y. | | Deposit date: | 2014-10-09 | | Release date: | 2015-02-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Evolutionarily Conserved Binding of Translationally Controlled Tumor Protein to Eukaryotic Elongation Factor 1B.
J.Biol.Chem., 290, 2015
|
|
2N51
 
 | |
2MVN
 
 | |
2P89
 
 | |
2QTF
 
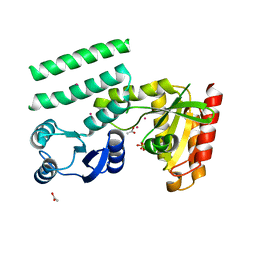 | | Crystal structure of a GTP-binding protein from the hyperthermophilic archaeon Sulfolobus solfataricus | | Descriptor: | ACETATE ION, CADMIUM ION, GTP-binding protein, ... | | Authors: | Wu, H, Sun, L, Brouns, S.J, Fu, S, Rao, Z, Van der Oost, J. | | Deposit date: | 2007-08-02 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a GTP-binding protein from the hyperthermophilic archaeon Sulfolobus solfataricus.
To be Published
|
|
2QTH
 
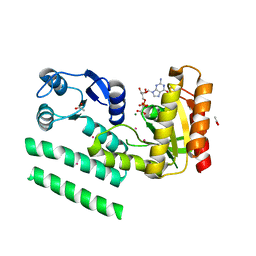 | | Crystal structure of a GTP-binding protein from the hyperthermophilic archaeon Sulfolobus solfataricus in complex with GDP | | Descriptor: | ACETATE ION, CADMIUM ION, GTP-binding protein, ... | | Authors: | Wu, H, Sun, L, Brouns, S.J, Fu, S, Rao, Z, Van der Oost, J. | | Deposit date: | 2007-08-02 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a GTP-binding protein from the hyperthermophilic archaeon Sulfolobus solfataricus.
To be Published
|
|
6IDE
 
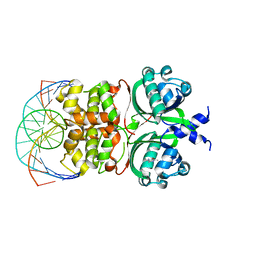 | | Crystal structure of the Vibrio cholera VqmA-Ligand-DNA complex provides molecular mechanisms for drug design | | Descriptor: | 3,5-dimethylpyrazin-2-ol, DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*AP*AP*TP*CP*CP*CP*CP*CP*CP*T)-3'), DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*TP*TP*TP*CP*CP*CP*CP*CP*CP*T)-3'), ... | | Authors: | Wu, H, Li, M.J, Guo, H.J, Zhou, H, Li, B, Xu, Q, Xu, C.Y, Yu, F, He, J.H. | | Deposit date: | 2018-09-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of theVibrio choleraeVqmA-ligand-DNA complex provides insight into ligand-binding mechanisms relevant for drug design.
J. Biol. Chem., 294, 2019
|
|
6KJU
 
 | | Huge conformation shift of Vibrio cholerae VqmA dimer in the absence of target DNA provides insight into DNA-binding mechanisms of LuxR-type receptors | | Descriptor: | 3,5-dimethylpyrazin-2-ol, Helix-turn-helix transcriptional regulator | | Authors: | Wu, H, Li, M.J, Guo, H.J, Zhou, H, Wang, W.W, Xu, Q, Xu, C.Y, Yu, F, He, J.H. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Large conformation shifts of Vibrio cholerae VqmA dimer in the absence of target DNA provide insight into DNA-binding mechanisms of LuxR-type receptors.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
4OR2
 
 | | Human class C G protein-coupled metabotropic glutamate receptor 1 in complex with a negative allosteric modulator | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-fluoro-N-methyl-N-{4-[6-(propan-2-ylamino)pyrimidin-4-yl]-1,3-thiazol-2-yl}benzamide, CHOLESTEROL, ... | | Authors: | Wu, H, Wang, C, Gregory, K.J, Han, G.W, Cho, H.P, Xia, Y, Niswender, C.M, Katritch, V, Cherezov, V, Conn, P.J, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2014-02-10 | | Release date: | 2014-03-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator
Science, 344, 2014
|
|
8ERT
 
 | | NLRP3 PYD filament | | Descriptor: | NACHT, LRR and PYD domains-containing protein 3 | | Authors: | Wu, H, Xiao, L. | | Deposit date: | 2022-10-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the active NLRP3 inflammasome disc.
Nature, 613, 2023
|
|
4QQK
 
 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) with GMS | | Descriptor: | (5S)-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}-N~6~-carbamimidoyl-L-lysine, GLYCEROL, Protein arginine N-methyltransferase 6, ... | | Authors: | Dong, A, Zeng, H, He, H, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
6SCX
 
 | | Crystal structure of the catalytic domain of human NUDT12 in complex with 7-methyl-guanosine-5'-triphosphate | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, CADMIUM ION, Peroxisomal NADH pyrophosphatase NUDT12 | | Authors: | McCarthy, A.A, Chen, K.M, Wu, H, Li, L, Homolka, D, Gos, P, Fleury-Olela, F, Pillai, R.S. | | Deposit date: | 2019-07-25 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Decapping Enzyme NUDT12 Partners with BLMH for Cytoplasmic Surveillance of NAD-Capped RNAs.
Cell Rep, 29, 2019
|
|
6TR3
 
 | | Ruminococcus gnavus GH29 fucosidase E1_10125 in complex with fucose | | Descriptor: | CALCIUM ION, F5/8 type C domain-containing protein, MAGNESIUM ION, ... | | Authors: | Owen, C.D, Wu, H, Crost, E, Colvile, A, Juge, N, Walsh, M.A. | | Deposit date: | 2019-12-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fucosidases from the human gut symbiont Ruminococcus gnavus.
Cell.Mol.Life Sci., 78, 2021
|
|
6TR4
 
 | | Ruminococcus gnavus GH29 fucosidase E1_10125 D221A mutant in complex with fucose | | Descriptor: | CALCIUM ION, CHLORIDE ION, F5/8 type C domain-containing protein, ... | | Authors: | Owen, C.D, Wu, H, Crost, E, Colvile, A, Juge, N, Walsh, M.A. | | Deposit date: | 2019-12-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Fucosidases from the human gut symbiont Ruminococcus gnavus.
Cell.Mol.Life Sci., 78, 2021
|
|
6DBR
 
 | | Cryo-EM structure of RAG in complex with one melted RSS and one unmelted RSS | | Descriptor: | CALCIUM ION, Forward strand of melted RSS substrate DNA, Forward strand of unmelted RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBL
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Molecule name: Forward strand of 12-RSS substrate DNA, Molecule name: Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (5.001 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBQ
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Molecule name: Forward strand of 12-RSS substrate DNA, Molecule name: Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBW
 
 | | Cryo-EM structure of RAG in complex with 12-RSS substrate DNA | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBT
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
