7NLW
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 2-(5-methoxy-1H-indol-3-yl)acetonitrile | | Descriptor: | 2-(5-methoxy-1~{H}-indol-3-yl)ethanenitrile, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLP
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with L-canavanine | | Descriptor: | Acetylglutamate kinase, L-CANAVANINE, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.213 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLX
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 7-(trifluoromethyl)quinolin-4-ol. | | Descriptor: | 7-(trifluoromethyl)quinolin-4-ol, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.234 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLF
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in apo form. | | Descriptor: | 1,2-ETHANEDIOL, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLU
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 1-(1H-indol-3-yl)ethan-1-one | | Descriptor: | 1-(1~{H}-indol-3-yl)ethanone, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.235 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NN7
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with dimethyl 5-hydroxyisophthalate. | | Descriptor: | 1,2-ETHANEDIOL, Acetylglutamate kinase, SULFATE ION, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.172 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NWR
 
 | | Structure of BT1526, a myo-inositol-1-phosphate synthase | | Descriptor: | Inositol-3-phosphate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Basle, A, Tang, G, Marles-Wright, J, Campopiano, D. | | Deposit date: | 2021-03-17 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of inositol lipid metabolism in gut-associated Bacteroidetes.
Nat Microbiol, 7, 2022
|
|
2VSH
 
 | | Synthesis of CDP-activated ribitol for teichoic acid precursors in Streptococcus pneumoniae | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Baur, S, Marles-Wright, J, Buckenmaier, S, Lewis, R.J, Vollmer, W. | | Deposit date: | 2008-04-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis of Cdp-Activated Ribitol for Teichoic Acid Precursors in Streptococcus Pneumoniae.
J.Bacteriol., 191, 2009
|
|
2VY9
 
 | | Molecular architecture of the stressosome, a signal integration and transduction hub | | Descriptor: | ANTI-SIGMA-FACTOR ANTAGONIST | | Authors: | Marles-Wright, J, Grant, T, Delumeau, O, van Duinen, G, Firbank, S.J, Lewis, P.J, Murray, J.W, Newman, J.A, Quin, M.B, Race, P.R, Rohou, A, Tichelaar, W, van Heel, M, Lewis, R.J. | | Deposit date: | 2008-07-21 | | Release date: | 2008-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Architecture of the "Stressosome," a Signal Integration and Transduction Hub
Science, 322, 2008
|
|
2VSI
 
 | | Synthesis of CDP-activated ribitol for teichoic acid precursors in Streptococcus pneumoniae | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, 4-AMINO-1-{5-O-[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]-ALPHA-D-ARABINOFURANOSYL}PYRIMIDIN-2(1H)-ONE, CALCIUM ION | | Authors: | Baur, S, Marles-Wright, J, Buckenmaier, S, Lewis, R.J, Vollmer, W. | | Deposit date: | 2008-04-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Synthesis of Cdp-Activated Ribitol for Teichoic Acid Precursors in Streptococcus Pneumoniae.
J.Bacteriol., 191, 2009
|
|
3ZT9
 
 | | The bacterial stressosome: a modular system that has been adapted to control secondary messenger signaling | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, SERINE PHOSPHATASE | | Authors: | Quin, M.B, Berrisford, J.M, Newman, J.A, Basle, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2011-07-06 | | Release date: | 2012-02-22 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Bacterial Stressosome: A Modular System that Has Been Adapted to Control Secondary Messenger Signaling.
Structure, 20, 2012
|
|
3ZTA
 
 | | The bacterial stressosome: a modular system that has been adapted to control secondary messenger signaling | | Descriptor: | ANTI-SIGMA-FACTOR ANTAGONIST (STAS) DOMAIN PROTEIN | | Authors: | Quin, M.B, Berrisford, J.M, Newman, J.A, Basle, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2011-07-06 | | Release date: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Bacterial Stressosome: A Modular System that Has Been Adapted to Control Secondary Messenger Signaling.
Structure, 20, 2012
|
|
3ZTB
 
 | | The bacterial stressosome: a modular system that has been adapted to control secondary messenger signaling | | Descriptor: | ANTI-SIGMA-FACTOR ANTAGONIST (STAS) DOMAIN PROTEIN, IODIDE ION | | Authors: | Quin, M.B, Berrisford, J.M, Newman, J.A, Basle, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2011-07-06 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Bacterial Stressosome: A Modular System that Has Been Adapted to Control Secondary Messenger Signaling.
Structure, 20, 2012
|
|
3ZXJ
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, HIAXHD3, ... | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZXK
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIAXHD3, alpha-L-arabinofuranose-(1-2)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZXN
 
 | | Moorella thermoacetica RsbS S58E | | Descriptor: | ANTI-SIGMA-FACTOR ANTAGONIST (STAS) DOMAIN PROTEIN, THIOCYANATE ION | | Authors: | Quin, M.B, Berrisford, J.M, Newman, J.A, Basle, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2011-08-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Bacterial Stressosome: A Modular System that Has Been Adapted to Control Secondary Messenger Signaling.
Structure, 20, 2012
|
|
3ZXL
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HIAXHD3 | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AXJ
 
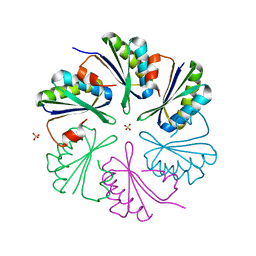 | | Structure of the Clostridium difficile EutM protein | | Descriptor: | ETHANOLAMINE CARBOXYSOME STRUCTURAL PROTEIN, SULFATE ION | | Authors: | Pitts, A.C, Tuck, L.R, Faulds-Pain, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2012-06-13 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Insight Into the Clostridium Difficile Ethanolamine Utilisation Microcompartment.
Plos One, 7, 2012
|
|
4AXI
 
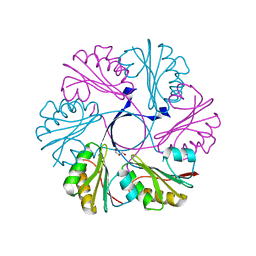 | | Structure of the Clostridium difficile EutS protein | | Descriptor: | ETHANOLAMINE CARBOXYSOME STRUCTURAL PROTEIN, GLYCEROL | | Authors: | Pitts, A.C, Tuck, L.R, Faulds-Pain, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2012-06-13 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural Insight Into the Clostridium Difficile Ethanolamine Utilisation Microcompartment.
Plos One, 7, 2012
|
|
4AXO
 
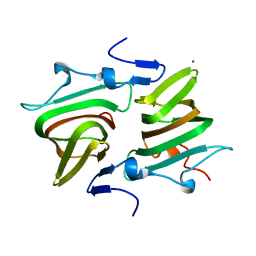 | | Structure of the Clostridium difficile EutQ protein | | Descriptor: | ETHANOLAMINE UTILIZATION PROTEIN, MAGNESIUM ION | | Authors: | Pitts, A.C, Tuck, L.R, Faulds-Pain, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2012-06-13 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural Insight Into the Clostridium Difficile Ethanolamine Utilisation Microcompartment.
Plos One, 7, 2012
|
|
4TQX
 
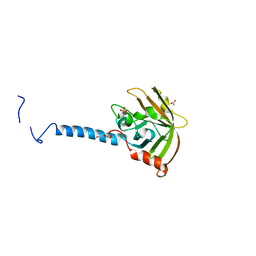 | | Molecular Basis of Streptococcus mutans Sortase A Inhibition by Chalcone. | | Descriptor: | ACETIC ACID, SULFATE ION, Sortase, ... | | Authors: | Wallock-Richards, D.J, Marles-Wright, J, Clarke, D.J, Maitra, A, Dodds, M, Hanley, B, Campopiano, D.J. | | Deposit date: | 2014-06-12 | | Release date: | 2015-05-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Molecular basis of Streptococcus mutans sortase A inhibition by the flavonoid natural product trans-chalcone.
Chem.Commun.(Camb.), 51, 2015
|
|
4C3S
 
 | | Structure of a propionaldehyde dehydrogenase from the Clostridium phytofermentans fucose utilisation bacterial microcompartment | | Descriptor: | ALDEHYDE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Marles-Wright, J, Crawshaw, A, Ang, T.F, Altenbach, K. | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Insight Into Coenzyme a Cofactor Binding and the Mechanism of Acyl-Transfer in an Acylating Aldehyde Dehydrogenase from Clostridium Phytofermentans.
Sci.Rep., 6, 2016
|
|
2CDG
 
 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide-specific T cell receptors (TCR 5B) | | Descriptor: | TCR 5E | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
2CDE
 
 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide specific T cell receptors - iNKT-TCR | | Descriptor: | INKT-TCR | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
2CDF
 
 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide-specific T cell receptors (TCR 5E) | | Descriptor: | TCR 5E | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
