5OT0
 
 | | The thermostable L-asparaginase from Thermococcus kodakarensis | | Descriptor: | 1,2-ETHANEDIOL, L-asparaginase, PHOSPHATE ION, ... | | Authors: | Guo, J, Coker, A.R, Wood, S.P, Cooper, J.B, Rashid, N, Chohan, S.M, Akhtar, M. | | Deposit date: | 2017-08-18 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure and function of the thermostable L-asparaginase from Thermococcus kodakarensis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5OV4
 
 | | Bacillus megaterium porphobilinogen deaminase D82A mutant | | Descriptor: | Porphobilinogen deaminase | | Authors: | Guo, J, Erskine, P, Coker, A.R, Wood, S.P, Cooper, J.B. | | Deposit date: | 2017-08-28 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Structural studies of domain movement in active-site mutants of porphobilinogen deaminase from Bacillus megaterium.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
2WQT
 
 | | Dodecahedral assembly of MhpD | | Descriptor: | 2-KETO-4-PENTENOATE HYDRATASE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Montgomery, M.G, Wood, S.P. | | Deposit date: | 2009-08-27 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Assembly of a 20Nm Protein Cage by Escherichia Coli 2-Hydroxypentadienoic Acid Hydratase (Mhpd).
J.Mol.Biol., 396, 2010
|
|
1YLV
 
 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH LAEVULINIC ACID | | Descriptor: | LAEVULINIC ACID, PROTEIN (5-AMINOLAEVULINIC ACID DEHYDRATASE), ZINC ION | | Authors: | Erskine, P.T, Newbold, R, Roper, J, Coker, A, Warren, M.J, Shoolingin-Jordan, P.M, Wood, S.P, Cooper, J.B. | | Deposit date: | 1999-02-22 | | Release date: | 2000-02-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Schiff base complex of yeast 5-aminolaevulinic acid dehydratase with laevulinic acid.
Protein Sci., 8, 1999
|
|
2C1H
 
 | | The X-ray Structure of Chlorobium vibrioforme 5-Aminolaevulinic Acid Dehydratase Complexed with a Diacid Inhibitor | | Descriptor: | 4,7-DIOXOSEBACIC ACID, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, MAGNESIUM ION | | Authors: | Coates, L, Beaven, G, Erskine, P.T, Beale, S, Wood, S.P, Shoolingin-Jordan, P.M, Cooper, J.B. | | Deposit date: | 2005-09-14 | | Release date: | 2005-12-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Chlorobium Vibrioforme 5-Aminolaevulinic Acid Dehydratase Complexed with a Diacid Inhibitor.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2CCM
 
 | | X-ray structure of Calexcitin from Loligo pealeii at 1.8A | | Descriptor: | CALCIUM ION, CALEXCITIN | | Authors: | Erskine, P.T, Beaven, G.D.E, Wood, S.P, Fox, G, Vernon, J, Giese, K.P, Cooper, J.B. | | Deposit date: | 2006-01-16 | | Release date: | 2006-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Neuronal Protein Calexcitin Suggests a Mode of Interaction in Signalling Pathways of Learning and Memory.
J.Mol.Biol., 357, 2006
|
|
1N28
 
 | | Crystal structure of the H48Q mutant of human group IIA phospholipase A2 | | Descriptor: | CALCIUM ION, Phospholipase A2, membrane associated | | Authors: | Edwards, S.H, Thompson, D, Baker, S.F, Wood, S.P, Wilton, D.C. | | Deposit date: | 2002-10-22 | | Release date: | 2003-10-28 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the H48Q active site mutant of human group IIA secreted phospholipase A2 at 1.5 A resolution provides an insight into the catalytic mechanism
Biochemistry, 41, 2002
|
|
1N29
 
 | | Crystal structure of the N1A mutant of human group IIA phospholipase A2 | | Descriptor: | CALCIUM ION, Phospholipase A2, membrane associated | | Authors: | Edwards, S.H, Thompson, D, Baker, S.F, Wood, S.P, Wilton, D.C. | | Deposit date: | 2002-10-22 | | Release date: | 2003-10-28 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of the H48Q active site mutant of human group IIA secreted phospholipase A2 at 1.5 A resolution provides an insight into the catalytic mechanism
Biochemistry, 41, 2002
|
|
1OD1
 
 | | Endothiapepsin PD135,040 complex | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide, SULFATE ION | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Cooper, J.B. | | Deposit date: | 2003-02-12 | | Release date: | 2003-06-12 | | Last modified: | 2012-11-30 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The Structure of Endothiapepsin Complexed with the Gem-Diol Inhibitor Pd-135,040 at 1.37 A
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4AVV
 
 | | Structure of CPHPC bound to Serum Amyloid P Component | | Descriptor: | (2R)-1-[6-[(2R)-2-carboxypyrrolidin-1-yl]-6-oxidanylidene-hexanoyl]pyrrolidine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kolstoe, S.E, Jenvey, M.C, Wood, S.P. | | Deposit date: | 2012-05-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interaction of Serum Amyloid P Component with Hexanoyl Bis(D-Proline) (Cphpc)
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AVS
 
 | |
4AVT
 
 | | Structure of CPHPC bound to Serum Amyloid P Component | | Descriptor: | (2R)-1-[6-[(2R)-2-carboxypyrrolidin-1-yl]-6-oxidanylidene-hexanoyl]pyrrolidine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kolstoe, S.E, Purvis, A, Wood, S.P. | | Deposit date: | 2012-05-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Interaction of Serum Amyloid P Component with Hexanoyl Bis(D-Proline) (Cphpc)
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AYU
 
 | | Structure of N-Acetyl-D-Proline bound to serum amyloid P component | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Hughes, P, Kolstoe, S.E, Wood, S.P. | | Deposit date: | 2012-06-22 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interaction of Serum Amyloid P Component with Hexanoyl Bis(D-Proline) (Cphpc)
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1OEW
 
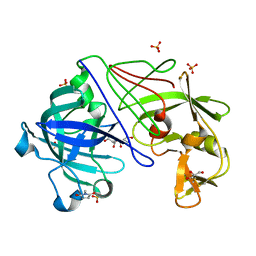 | | ATOMIC RESOLUTION STRUCTURE OF NATIVE ENDOTHIAPEPSIN | | Descriptor: | ENDOTHIAPEPSIN, GLYCEROL, SERINE, ... | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2003-03-31 | | Release date: | 2003-04-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
1OEX
 
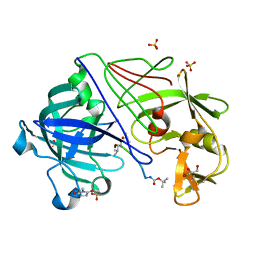 | | Atomic Resolution Structure of Endothiapepsin in Complex with a Hydroxyethylene Transition State Analogue Inhibitor H261 | | Descriptor: | ENDOTHIAPEPSIN, GLYCEROL, INHIBITOR H261, ... | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2003-03-31 | | Release date: | 2003-04-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
1OHL
 
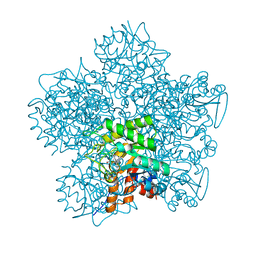 | | YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE PUTATIVE CYCLIC REACTION INTERMEDIATE COMPLEX | | Descriptor: | 3-[5-(AMINOMETHYL)-4-(CARBOXYMETHYL)-1H-PYRROL-3-YL]PROPANOIC ACID, BETA-MERCAPTOETHANOL, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, ... | | Authors: | Erskine, P.T, Coates, L, Butler, D, Youell, J.H, Brindley, A.A, Wood, S.P, Warren, M.J, Shoolingin-Jordan, P.M, Cooper, J.B. | | Deposit date: | 2003-05-27 | | Release date: | 2003-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-Ray Structure of a Putative Reaction Intermediateof 5-Aminolaevulinic Acid Dehydratase
Biochem.J., 373, 2003
|
|
1PDA
 
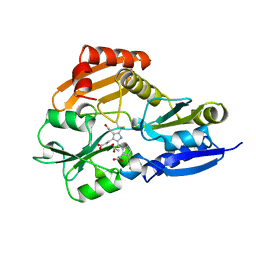 | | STRUCTURE OF PORPHOBILINOGEN DEAMINASE REVEALS A FLEXIBLE MULTIDOMAIN POLYMERASE WITH A SINGLE CATALYTIC SITE | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, ACETIC ACID, PORPHOBILINOGEN DEAMINASE | | Authors: | Louie, G.V, Brownlie, P.D, Lambert, R, Cooper, J.B, Blundell, T.L, Wood, S.P, Warren, M.J, Woodcock, S.C, Jordan, P.M. | | Deposit date: | 1992-11-17 | | Release date: | 1993-10-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of porphobilinogen deaminase reveals a flexible multidomain polymerase with a single catalytic site.
Nature, 359, 1992
|
|
1QN2
 
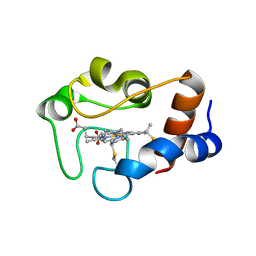 | | cytochrome cH from Methylobacterium extorquens | | Descriptor: | CYTOCHROME CH, HEME C | | Authors: | Read, J, Gill, R, Dales, S.L, Cooper, J.B, Wood, S.P, Anthony, C. | | Deposit date: | 1999-10-13 | | Release date: | 2000-10-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Molecular Structure of an Unusual Cytochrome C2 Determined at 2.0A; the Cytochrome cH from Methylobacterium Extorquens
Protein Sci., 8, 1999
|
|
4EGH
 
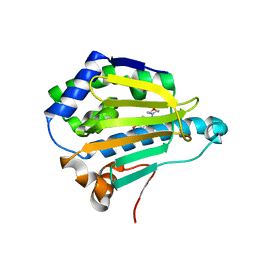 | |
4EGK
 
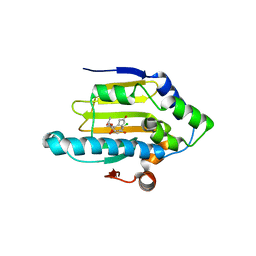 | | Human Hsp90-alpha ATPase domain bound to Radicicol | | Descriptor: | Heat shock protein HSP 90-alpha, RADICICOL | | Authors: | Chen, W.J, Wood, S.P. | | Deposit date: | 2012-03-31 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Fragment screening using capillary electrophoresis (CEfrag) for hit identification of heat shock protein 90 ATPase inhibitors.
J Biomol Screen, 17, 2012
|
|
4EGI
 
 | | Hsp90-alpha ATPase domain in complex with 2-Amino-4-ethylthio-6-methyl-1,3,5-triazine | | Descriptor: | 4-(ethylsulfanyl)-6-methyl-1,3,5-triazin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Chen, W.J, Wood, S.P. | | Deposit date: | 2012-03-31 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment screening using capillary electrophoresis (CEfrag) for hit identification of heat shock protein 90 ATPase inhibitors.
J Biomol Screen, 17, 2012
|
|
1EB3
 
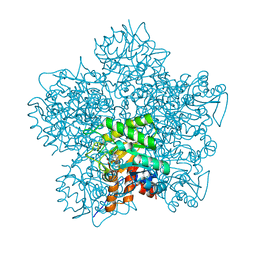 | | YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE 4,7-DIOXOSEBACIC ACID COMPLEX | | Descriptor: | 4,7-DIOXOSEBACIC ACID, 5-AMINOLAEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Coates, L, Newbold, R, Brindley, A.A, Stauffer, F, Wood, S.P, Warren, M.J, Cooper, J.B, Shoolingin-Jordan, P.M, Neier, R. | | Deposit date: | 2001-07-18 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Two Diacid Inhibitors
FEBS Lett., 503, 2001
|
|
4HTG
 
 | | Porphobilinogen Deaminase from Arabidopsis Thaliana | | Descriptor: | 3-[(5Z)-5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methylidene}-4-(carboxymethyl)-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]propanoic acid, ACETATE ION, Porphobilinogen deaminase, ... | | Authors: | Roberts, A, Gill, R, Hussey, R.J, Erskine, P.T, Cooper, J.B, Wood, S.P, Chrystal, E.J.T, Shoolingin-Jordan, P.M. | | Deposit date: | 2012-11-01 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into the mechanism of pyrrole polymerization catalysed by porphobilinogen deaminase: high-resolution X-ray studies of the Arabidopsis thaliana enzyme.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1GN6
 
 | | G152A mutant of Mycobacterium tuberculosis iron-superoxide dismutase. | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE | | Authors: | Bunting, K.A, Cooper, J.B, Saward, S, Erskine, P.T, Badasso, M.O, Wood, S.P, Zhang, Y, Young, D.B. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-05 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-Ray Structure Analysis of an Engineered Fe-Superoxide Dismutase Gly-Ala Mutant with Significantly Reduced Stability to Denaturant
FEBS Lett., 387, 1996
|
|
1GKT
 
 | | Neutron Laue diffraction structure of endothiapepsin complexed with transition state analogue inhibitor H261 | | Descriptor: | ENDOTHIAPEPSIN, INHIBITOR, H261 | | Authors: | Coates, L, Erskine, P.T, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2001-08-20 | | Release date: | 2001-11-20 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | A Neutron Laue Diffraction Study of Endothiapepsin: Implications for the Aspartic Proteinase Mechanism
Biochemistry, 40, 2001
|
|
